A Metagenomic Analysis Provides a Culture-Independent Pathogen Detection for Atopic Dermatitis
- Affiliations
-
- 1Department of Internal Medicine, Ewha Womans University School of Medicine, Seoul, Korea.
- 2Department of Computer Science and Engineering, Hanyang University, Seoul, Korea.
- 3Asan Institute for Life Sciences, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 4Division of Molecular and Life Sciences, Department of Life Science, Pohang University of Science and Technology, Pohang, Korea.
- 5Department of Allergy and Clinical Immunology, Ajou University Medical Center, Suwon, Korea.
- 6Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea.
- 7Department of Pediatrics, Pediatric Allergy and Respiratory Center, Soonchunhyang University Hospital, Soonchunhyang University College of Medicine, Seoul, Korea. bypyun@schmc.ac.kr
- 8Institute of MD Healthcare, Seoul, Korea. arraychip@gmail.com
- KMID: 2383994
- DOI: http://doi.org/10.4168/aair.2017.9.5.453
Abstract
- PURPOSE
Atopic dermatitis (AD) is an inflammatory skin disease, significantly affecting the quality of life. Using AD as a model system, we tested a successive identification of AD-associated microbes, followed by a culture-independent serum detection of the identified microbe.
METHODS
A total of 43 genomic DNA preparations from washing fluid of the cubital fossa of 6 healthy controls, skin lesions of 27 AD patients, 10 of which later received treatment (post-treatment), were subjected to high-throughput pyrosequencing on a Roche 454 GS-FLX platform.
RESULTS
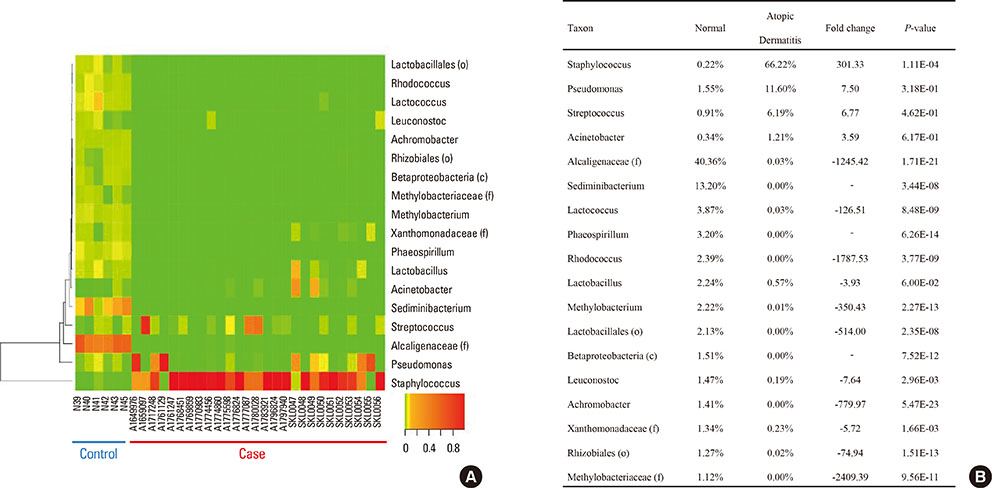
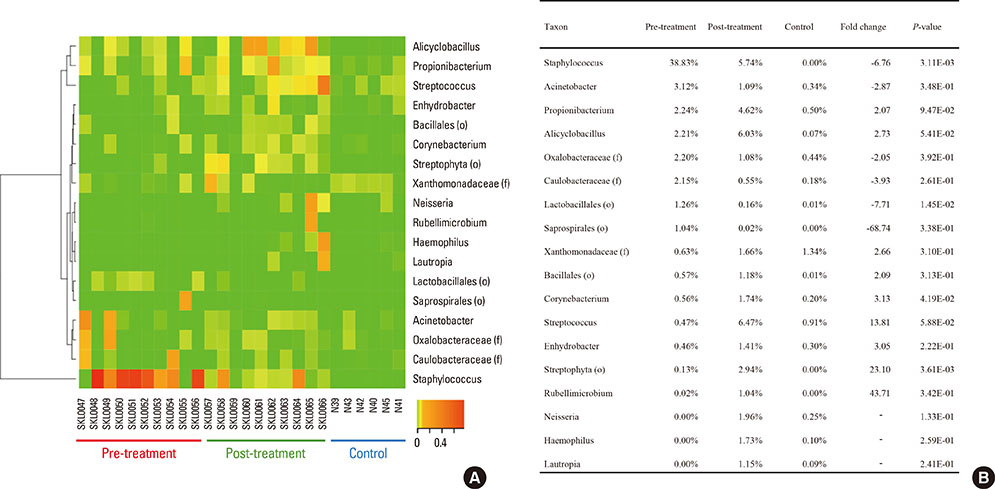
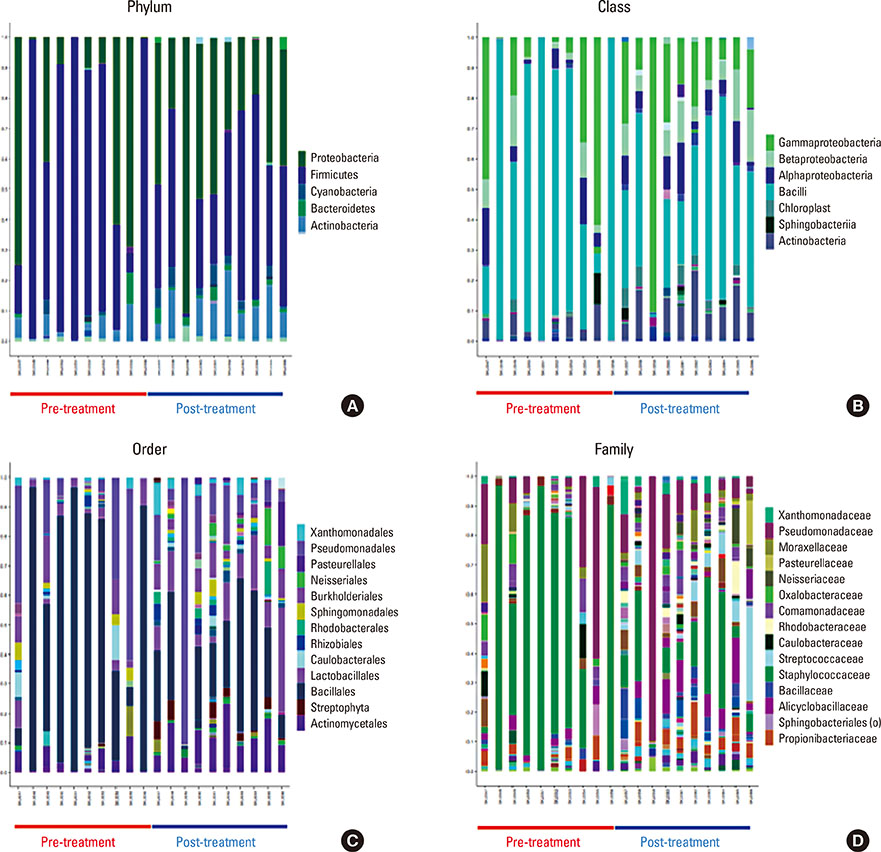
Microbial diversity was decreased in AD, and was restored following treatment. AD was characterized by the domination of Staphylococcus, Pseudomonas, and Streptococcus, whereas Alcaligenaceae (f), Sediminibacterium, and Lactococcus were characteristic of healthy skin. An enzyme-linked immunosorbent assay (ELISA) showed that serum could be used as a source for the detection of Staphylococcus aureus extracellular vesicles (EVs). S. aureus EV-specific immunoglobulin G (IgG) and immunoglobulin E (IgE) were quantified in the serum.
CONCLUSIONS
A metagenomic analysis together with a serum detection of pathogen-specific EVs provides a model for successive identification and diagnosis of pathogens of AD.
MeSH Terms
-
Alcaligenaceae
Dermatitis, Atopic*
Diagnosis
DNA
Enzyme-Linked Immunosorbent Assay
Extracellular Vesicles
Humans
Immunoglobulin E
Immunoglobulin G
Immunoglobulins
Lactococcus
Metagenomics*
Pseudomonas
Quality of Life
Skin
Skin Diseases
Staphylococcus
Staphylococcus aureus
Streptococcus
DNA
Immunoglobulin E
Immunoglobulin G
Immunoglobulins
Figure
Cited by 4 articles
-
Microbiome Research in Atopic Dermatitis
Min-Hye Kim
Hanyang Med Rev. 2018;38(2):85-92. doi: 10.7599/hmr.2018.38.2.85.Lactobacillus plantarum-derived Extracellular Vesicles Protect Atopic Dermatitis Induced by Staphylococcus aureus-derived Extracellular Vesicles
Min-Hye Kim, Seng Jin Choi, Hyun-Il Choi, Jun-Pyo Choi, Han-Ki Park, Eun Kyoung Kim, Min-Jeong Kim, Byoung Seok Moon, Taek-ki Min, Mina Rho, Young-Joo Cho, Sanghwa Yang, Yoon-Keun Kim, You-Young Kim, Bok Yang Pyun
Allergy Asthma Immunol Res. 2018;10(5):516-532. doi: 10.4168/aair.2018.10.5.516.Current research status of pediatric atopic dermatitis in Korea
Bok Yang Pyun
Allergy Asthma Respir Dis. 2018;6(Suppl 1):S40-S43. doi: 10.4168/aard.2018.6.S1.S40.Microbiome in the Gut-Skin Axis in Atopic Dermatitis
So-Yeon Lee, Eun Lee, Yoon Mee Park, Soo-Jong Hong
Allergy Asthma Immunol Res. 2018;10(4):354-362. doi: 10.4168/aair.2018.10.4.354.
Reference
-
1. Sabin BR, Peters N, Peters AT. Chapter 20: atopic dermatitis. Allergy Asthma Proc. 2012; 33:Suppl 1. S67–S69.2. Lee JH, Son SW, Cho SH. A comprehensive review of the treatment of atopic eczema. Allergy Asthma Immunol Res. 2016; 8:181–190.3. Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004; 113:651–657.4. Pyun BY. Natural history and risk factors of atopic dermatitis in children. Allergy Asthma Immunol Res. 2015; 7:101–105.5. Bieber T. Atopic dermatitis. N Engl J Med. 2008; 358:1483–1494.6. Ahn K. The prevalence of atopic dermatitis in Korean children. Allergy Asthma Immunol Res. 2016; 8:1–2.7. Lee JH, Han KD, Kim KM, Park YG, Lee JY, Park YM. Prevalence of atopic dermatitis in Korean children based on data from the 2008–2011 Korean National Health and Nutrition Examination Survey. Allergy Asthma Immunol Res. 2016; 8:79–83.8. Niebuhr M, Langnickel J, Draing C, Renz H, Kapp A, Werfel T. Dysregulation of toll-like receptor-2 (TLR-2)-induced effects in monocytes from patients with atopic dermatitis: impact of the TLR-2 R753Q polymorphism. Allergy. 2008; 63:728–734.9. Wollenberg A, Feichtner K. Atopic dermatitis and skin allergies - update and outlook. Allergy. 2013; 68:1509–1519.10. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012; 22:850–859.11. Breuer K, Wittmann M, Bösche B, Kapp A, Werfel T. Severe atopic dermatitis is associated with sensitization to staphylococcal enterotoxin B (SEB). Allergy. 2000; 55:551–555.12. Chen YE, Tsao H. The skin microbiome: current perspectives and future challenges. J Am Acad Dermatol. 2013; 69:143–155.13. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214.14. EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013; 12:347–357.15. Pyun BY. Extracellular vesicle: an unknown environmental factor for causing airway disease. Allergy Asthma Immunol Res. 2016; 8:179–180.16. Shin TS, Kim JH, Kim YS, Jeon SG, Zhu Z, Gho YS, et al. Extracellular vesicles are key intercellular mediators in the development of immune dysfunction to allergens in the airways. Allergy. 2010; 65:1256–1265.17. Hong SW, Kim MR, Lee EY, Kim JH, Kim YS, Jeon SG, et al. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 2011; 66:351–359.18. Hong SW, Choi EB, Min TK, Kim JH, Kim MH, Jeon SG, et al. An important role of α-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS One. 2014; 9:e100499.19. Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol Suppl (Stockh). 1980; 92:44–47.20. Severity scoring of atopic dermatitis: the SCORAD index. Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology. 1993; 186:23–31.21. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461.22. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7:335–336.23. Drago L, De Grandi R, Altomare G, Pigatto P, Rossi O, Toscano M. Skin microbiota of first cousins affected by psoriasis and atopic dermatitis. Clin Mol Allergy. 2016; 14:2.24. Seite S, Flores GE, Henley JB, Martin R, Zelenkova H, Aguilar L, et al. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J Drugs Dermatol. 2014; 13:1365–1372.25. Choi Y, Park H, Park HS, Kim YK. Extracellular vesicles, a key mediator to link environmental microbiota to airway immunity. Allergy Asthma Immunol Res. 2017; 9:101–106.26. Kim JH, Lee J, Park J, Gho YS. Gram-negative and gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol. 2015; 40:97–104.27. Campos JH, Soares RP, Ribeiro K, Andrade AC, Batista WL, Torrecilhas AC. Extracellular vesicles: role in inflammatory responses and potential uses in vaccination in cancer and infectious diseases. J Immunol Res. 2015; 2015:832057.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Measurement of Atopic Dermatitis Disability
- Serum IgE Level in Patients of Atopic Dermatitis and Atopic Dermatitis with Molluscum Contagiosum
- Nipple Involvement in Atopic Dermatitis: Report of 3 cases
- A Case of Head and Neck Dermatitis Treated with Itraconazole
- Two Cases of Atopic Dermatitis Developing Ocular Complication and Immunological Disturbance