Ann Surg Treat Res.
2017 May;92(5):331-339. 10.4174/astr.2017.92.5.331.
Next-generation sequencing of BRCA1/2 in breast cancer patients: potential effects on clinical decision-making using rapid, high-accuracy genetic results
- Affiliations
-
- 1Department of Surgery, Yonsei University College of Medicine, Seoul, Korea. hyungseokpark.md@gmail.com
- 2Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 3Department of Internal Medicine, Yonsei University College of Medicine, Seoul, Korea.
- 4Brain Korea 21 PLUS Project for Medical Science, Yonsei University, Seoul, Korea.
- KMID: 2377703
- DOI: http://doi.org/10.4174/astr.2017.92.5.331
Abstract
- PURPOSE
We evaluated the clinical role of rapid next-generation sequencing (NGS) for identifying BRCA1/2 mutations compared to traditional Sanger sequencing.
METHODS
Twenty-four paired samples from 12 patients were analyzed in this prospective study to compare the performance of NGS to the Sanger method. Both NGS and Sanger sequencing were performed in 2 different laboratories using blood samples from patients with breast cancer. We then analyzed the accuracy of NGS in terms of variant calling and determining concordance rates of BRCA1/2 mutation detection.
RESULTS
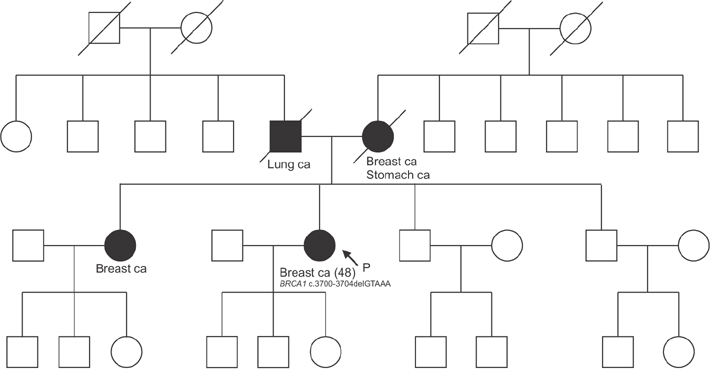
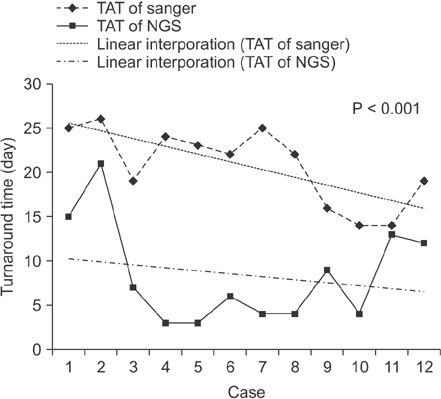
The overall concordance rate of BRCA1/2 mutation identification was 100%. Variants of unknown significance (VUS) were reported in two cases of BRCA1 and 3 cases of BRCA2 after Sanger sequencing, whereas NGS reported only 1 case of BRCA1 VUS, likely due to differences in reference databases used for mutation identification. The median turnaround time of Sanger sequencing was 22 days (range, 14-26 days), while the median time of NGS was only 6 days (range, 3-21 days).
CONCLUSION
NGS yielded comparably accurate results to Sanger sequencing and in a much shorter time with respect to BRCA1/2 mutation identification. The shorter turnaround time and higher accuracy of NGS may help clinicians make more timely and informed decisions regarding surgery or neoadjuvant chemotherapy in patients with breast cancer.
MeSH Terms
Figure
Reference
-
1. King MC, Marks JH, Mandell JB; New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003; 302:643–646.2. Schenberg T, Mitchell G. Prophylactic bilateral salpingectomy as a prevention strategy in women at high-risk of ovarian cancer: a mini-review. Front Oncol. 2014; 4:21.3. Wevers MR, Aaronson NK, Verhoef S, Bleiker EM, Hahn DE, Kuenen MA, et al. Impact of rapid genetic counselling and testing on the decision to undergo immediate or delayed prophylactic mastectomy in newly diagnosed breast cancer patients: findings from a randomised controlled trial. Br J Cancer. 2014; 110:1081–1087.4. Evans DG, Lalloo F, Ashcroft L, Shenton A, Clancy T, Baildam AD, et al. Uptake of risk-reducing surgery in unaffected women at high risk of breast and ovarian cancer is risk, age, and time dependent. Cancer Epidemiol Biomarkers Prev. 2009; 18:2318–2324.5. Schwartz MD, Lerman C, Brogan B, Peshkin BN, Halbert CH, DeMarco T, et al. Impact of BRCA1/BRCA2 counseling and testing on newly diagnosed breast cancer patients. J Clin Oncol. 2004; 22:1823–1829.6. Zanna I, Rizzolo P, Sera F, Falchetti M, Aretini P, Giannini G, et al. The BRCAPRO 5.0 model is a useful tool in genetic counseling and clinical management of male breast cancer cases. Eur J Hum Genet. 2010; 18:856–858.7. Metzker ML. Sequencing technologies: the next generation. Nat Rev Genet. 2010; 11:31–46.8. Chan M, Ji SM, Yeo ZX, Gan L, Yap E, Yap YS, et al. Development of a next-generation sequencing method for BRCA mutation screening: a comparison between a high-throughput and a benchtop platform. J Mol Diagn. 2012; 14:602–612.9. Han SA, Park SK, Ahn SH, Lee MH, Noh DY, Kim LS, et al. The Korean Hereditary Breast Cancer (KOHBRA) study: protocols and interim report. Clin Oncol (R Coll Radiol). 2011; 23:434–441.10. Friedman LS, Gayther SA, Kurosaki T, Gordon D, Noble B, Casey G, et al. Mutation analysis of BRCA1 and BRCA2 in a male breast cancer population. Am J Hum Genet. 1997; 60:313–319.11. Feliubadalo L, Lopez-Doriga A, Castellsague E, del Valle J, Menendez M, Tornero E, et al. Next-generation sequencing meets genetic diagnostics: development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur J Hum Genet. 2013; 21:864–870.12. Ozcelik H, Shi X, Chang MC, Tram E, Vlasschaert M, Di Nicola N, et al. Long-range PCR and next-generation sequencing of BRCA1 and BRCA2 in breast cancer. J Mol Diagn. 2012; 14:467–475.13. Bosdet IE, Docking TR, Butterfield YS, Mungall AJ, Zeng T, Coope RJ, et al. A clinically validated diagnostic secondgeneration sequencing assay for detection of hereditary BRCA1 and BRCA2 mutations. J Mol Diagn. 2013; 15:796–809.14. Michils G, Hollants S, Dehaspe L, Van Houdt J, Bidet Y, Uhrhammer N, et al. Molecular analysis of the breast cancer genes BRCA1 and BRCA2 using amplicon-based massive parallel pyrosequencing. J Mol Diagn. 2012; 14:623–630.15. Hernan I, Borras E, de Sousa Dias M, Gamundi MJ, Mane B, Llort G, et al. Detection of genomic variations in BRCA1 and BRCA2 genes by long-range PCR and next-generation sequencing. J Mol Diagn. 2012; 14:286–293.16. National Comprehensive Cancer Network. NCCN guidelines Version 3.2014. NCCN guidelines for treatment of cancer by site: breast cancer [Internet]. Fort Wathington (PA): National Comprehensive Cancer Network;c2017. cited 2014 Oct 1. Available from: https://www.nccn.org/professionals/physician_gls/f_guidelines.asp#site.17. Arrington AK, Jarosek SL, Virnig BA, Habermann EB, Tuttle TM. Patient and surgeon characteristics associated with increased use of contralateral prophylactic mastectomy in patients with breast cancer. Ann Surg Oncol. 2009; 16:2697–2704.18. Association for Molecular Pathology v. Myriad Genetics, Inc. [Internet]. SCOTUblog;c2017. cited 2017 Apr 3. Available from: http://www.scotusblog.com/case-files/cases/association-for-molecular-pathology-v-myriad-genetics-inc./.19. Tucker KI. Genetics lab refuses to share data that could save lives; why myriad genetics hoards its information about BRCA mutations [Internet]. New York: The Forward Association Inc;c2017. cited 2014 Sep 27. Available from: http://forward.com/culture/203739/genetics-labrefuses-to-share-data-that-could-save/.20. So D, Joly Y. Commercial opportunities and ethical pitfalls in personalized medicine: a Myriad of reasons to revisit the Myriad Genetics Saga. Curr Pharmacogenomics Person Med. 2013; 11:98–109.21. Nelson B. Prometheus bound, but myriad loose ends: amid new legal battles over BRCA tests, technology may resolve what the courts have not. Cancer Cytopathol. 2013; 121:535–536.22. Domchek SM, Greenberg RA. Breast cancer gene variants: separating the harmful from the harmless. J Clin Invest. 2009; 119:2895–2897.23. Richter S, Haroun I, Graham TC, Eisen A, Kiss A, Warner E. Variants of unknown significance in BRCA testing: impact on risk perception, worry, prevention and counseling. Ann Oncol. 2013; 24:Suppl 8. viii69–viii74.24. Murray ML, Cerrato F, Bennett RL, Jarvik GP. Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med. 2011; 13:998–1005.25. Moghadasi S, Hofland N, Wouts JN, Hogervorst FB, Wijnen JT, Vreeswijk MP, et al. Variants of uncertain significance in BRCA1 and BRCA2 assessment of in silico analysis and a proposal for communication in genetic counselling. J Med Genet. 2013; 50:74–79.26. Lu JT, Campeau PM, Lee BH. Genotype-phenotype correlation: promiscuity in the era of next-generation sequencing. N Engl J Med. 2014; 371:593–596.27. Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014; 156:872–877.28. Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014; 32:2001–2009.29. Robson M. Multigene panel testing: planning the next generation of research studies in clinical cancer genetics. J Clin Oncol. 2014; 32:1987–1989.30. Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011; 125:325–349.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Performance Evaluation of BRCA1/2 Genetic Test Using Next-Generation Sequencing Based on Target Capture Method
- Utility of Next-Generation Sequencing Panel Including Hereditary Breast and Ovarian Cancer-Related Genes for Pathogenic Variant Detection
- Evaluation of a Targeted Next-generation Sequencing Assay for BRCA Mutation Screening in Clinical Samples
- Clinical Implications of Genetic Testing for Hereditary Breast and Ovarian Cancer Syndrome in the Era of Genomic Medicine: Clinician's Perspectives
- Impact of Early Testing and Analysis of Germline Genetic Mutation in Patients with Breast Cancer: A Single Institution Experience