Lab Med Online.
2021 Oct;11(4):283-289. 10.47429/lmo.2021.11.4.283.
Evaluation of a Targeted Next-generation Sequencing Assay for BRCA Mutation Screening in Clinical Samples
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea
- 2Research & Development Centre, NGeneBio Co., Ltd., Seoul, Korea
- KMID: 2526090
- DOI: http://doi.org/10.47429/lmo.2021.11.4.283
Abstract
- Background
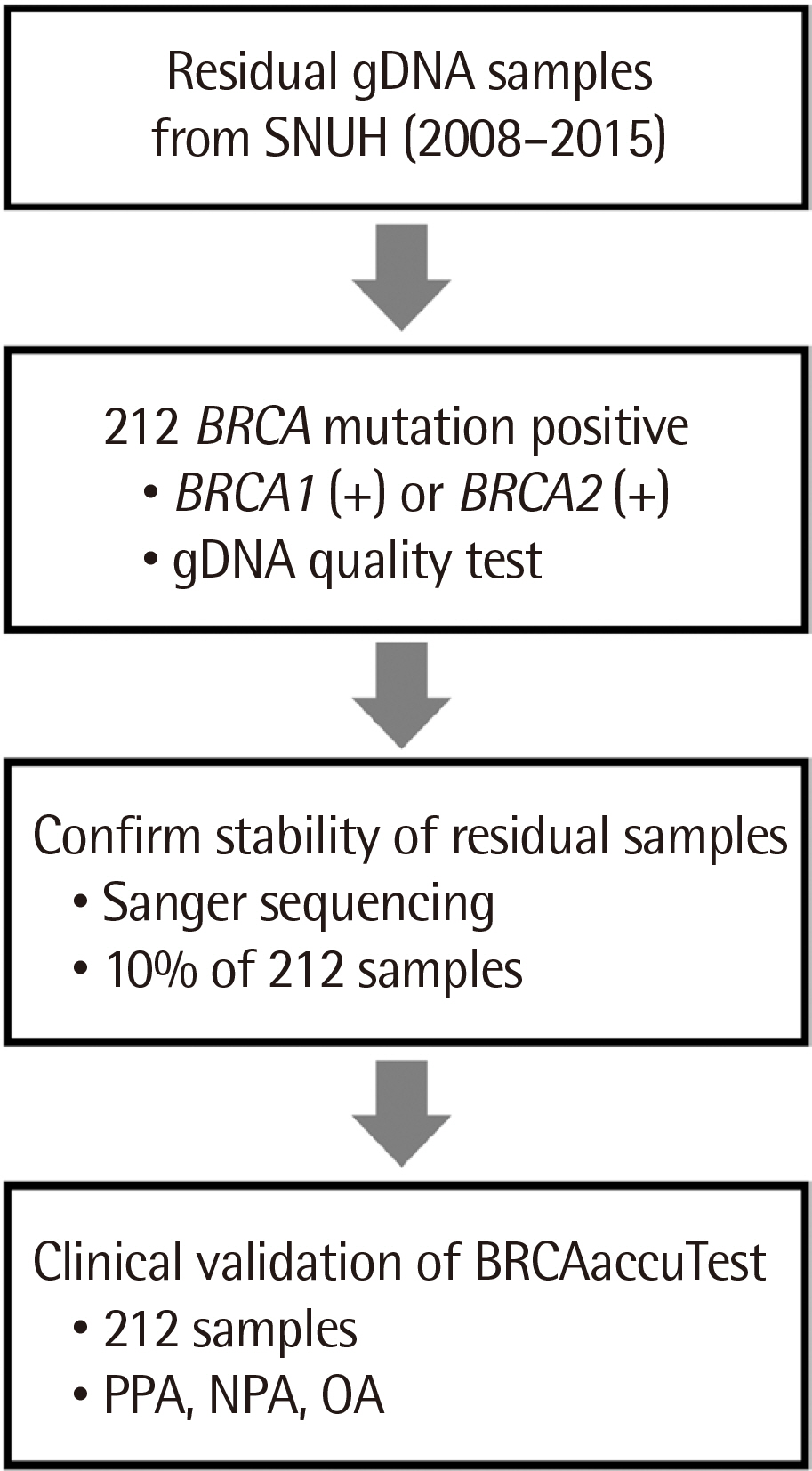
While several factors contribute to breast cancer pathogenesis, hereditary breast cancer results from a genetic predisposition. Genes associated with hereditary breast cancer may be divided into high- and low-penetrance genes depending on their risk rates. BRCA1 and BRCA2 are typical high-penetrance genes that increase the risk of developing breast and ovarian cancers upon undergoing mutations. This study aimed to evaluate the clinical performance of BRCAaccuTestTM (NGeneBio, Korea).
Methods
BRCAaccuTestTM is a reagent used to produce libraries for analyzing BRCA1/2 genes using next-generation sequencing (NGS), which analyzes blood-derived genomic DNA. Libraries with adapters and barcodes compatible with the Illumina platform were produced. The clinical performance of NGS-based BRCAaccuTestTM in identifying BRCA1/2mutations was compared with that of the traditional Sanger sequencing method. Both NGS and Sanger sequencing were performed in a single laboratory using archival DNA from blood samples of 212 patients with breast cancer.
Results
All target regions amplified were successfully sequenced to obtain a minimum coverage of 20, demonstrating 100% concordance with the pathogenic single-nucleotide variations and small insertions-deletions previously identified by Sanger sequencing.
Conclusions
This study demonstrates the feasibility of using BRCAaccuTestTM to detect the BRCA1/2 mutations with high accuracy.
Keyword
Figure
Reference
-
1. Heymach J, Krilov L, Alberg A, Baxter N, Chang SM, Corcoran RB, et al. 2018; Clinical Cancer Advances 2018: Annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol. 36:1020–44. DOI: 10.1200/JCO.2017.77.0446. PMID: 29380678.
Article2. Jung KW, Won YJ, Kong HJ, Lee ES. 2018; Prediction of cancer incidence and mortality in Korea, 2018. Cancer Res Treat. 50:317–23. DOI: 10.4143/crt.2018.142. PMID: 29566480. PMCID: PMC5912149.
Article3. Jung KW, Won YJ, Kong HJ, Lee ES. Community of Population-Based Regional Cancer R. 2018; Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2015. Cancer Res Treat. 50:303–16. DOI: 10.4143/crt.2018.143. PMID: 29566481. PMCID: PMC5912151.
Article4. Park HS, Park SJ, Kim JY, Kim S, Ryu J, Sohn J, et al. 2017; Next-generation sequencing of BRCA1/2 in breast cancer patients: potential effects on clinical decision-making using rapid, high-accuracy genetic results. Ann Surg Treat Res. 92:331–9. DOI: 10.4174/astr.2017.92.5.331. PMID: 28480178. PMCID: PMC5416916.5. Janavičius R. 2010; Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J. 1:397–412. DOI: 10.1007/s13167-010-0037-y. PMID: 23199084. PMCID: PMC3405339.6. Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. 2003; Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 72:1117–30. DOI: 10.1086/375033. PMID: 12677558. PMCID: PMC1180265.7. Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, et al. 2013; Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst. 105:812–22. DOI: 10.1093/jnci/djt095. PMID: 23628597.8. D'Argenio V, Esposito MV, Telese A, Precone V, Starnone F, Nunziato M, et al. 2015; The molecular analysis of BRCA1 and BRCA2: Next-generation sequencing supersedes conventional approaches. Clin Chim Acta. 446:221–5. DOI: 10.1016/j.cca.2015.03.045. PMID: 25896959.9. Wallace AJ. 2016; New challenges for BRCA testing: a view from the diagnostic laboratory. Eur J Hum Genet. 24 Suppl 1:S10–8. DOI: 10.1038/ejhg.2016.94. PMID: 27514839. PMCID: PMC5141576.10. Ruiz A, Llort G, Yagüe C, Baena N, Viñas M, Torra M, et al. 2014; Genetic testing in hereditary breast and ovarian cancer using massive parallel sequencing. Biomed Res Int. 2014:542541. DOI: 10.1155/2014/542541. PMID: 25136594. PMCID: PMC4098986.
Article11. Hajian-Tilaki K. 2014; Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform. 48:193–204. DOI: 10.1016/j.jbi.2014.02.013. PMID: 24582925.
Article12. Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ les (Version 1.33) [Software]. https://github.com/najoshi/sickle. Updated on Jul 2014.13. Li H, Durbin R. 2009; Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–60. DOI: 10.1093/bioinformatics/btp324. PMID: 19451168. PMCID: PMC2705234.
Article14. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. 2011; A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–8. DOI: 10.1038/ng.806. PMID: 21478889. PMCID: PMC3083463.
Article15. Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. https://arxiv.org/abs/1207.3907. Updated on Jul 2012.16. Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. 2012; A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92. DOI: 10.4161/fly.19695. PMID: 22728672. PMCID: PMC3679285.17. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article18. Kechin A, Khrapov E, Boyarskikh U, Kel A, Filipenko M. 2018; BRCA-analyzer: Automatic work ow for processing NGS reads of BRCA1 and BRCA2 genes. Comput Biol Chem. 77:297–306. DOI: 10.1016/j.compbiolchem.2018.10.012. PMID: 30408727.19. Ermolenko NA, Boyarskikh UA, Kechin AA, Mazitova AM, Khrapov EA, Petrova VD, et al. 2015; Massive parallel sequencing for diagnostic genetic testing of BRCA genes--a single center experience. Asian Pac J Cancer Prev. 16:7935–41. DOI: 10.7314/APJCP.2015.16.17.7935. PMID: 26625824.20. Suryavanshi M, Kumar D, Panigrahi MK, Chowdhary M, Mehta A. 2017; Detection of false positive mutations in BRCA gene by next generation sequencing. Fam Cancer. 16:311–7. DOI: 10.1007/s10689-016-9955-8. PMID: 27848044.21. Strom CM, Rivera S, Elzinga C, Angeloni T, Rosenthal SH, Goos-Root D, et al. 2015; Development and validation of a next-generation sequencing assay for BRCA1 and BRCA2 variants for the clinical laboratory. PLoS One. 10:e0136419. DOI: 10.1371/journal.pone.0136419. PMID: 26295337. PMCID: PMC4546651.22. Capone GL, Putignano AL, Trujillo Saavedra S, Paganini I, Sestini R, Gensini F, et al. 2018; Evaluation of a next-generation sequencing assay for BRCA1 and BRCA2 mutation detection. J Mol Diagn. 20:87–94. DOI: 10.1016/j.jmoldx.2017.09.005. PMID: 29061375.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- New Lung Cancer Panel for High-Throughput Targeted Resequencing
- Next generation sequencing and anti-cancer therapy
- Colorectal cancer with a germline BRCA1 variant inherited paternally: a case report
- The Incidence of Occult Malignancy in Contralateral Risk Reducing Mastectomy Among Affected Breast Cancer Gene Mutation Carriers in South Korea
- Exome sequencing in a breast cancer family without BRCA mutation