J Gastric Cancer.
2015 Mar;15(1):39-45. 10.5230/jgc.2015.15.1.39.
Loss of FAT Atypical Cadherin 4 Expression Is Associated with High Pathologic T Stage in Radically Resected Gastric Cancer
- Affiliations
-
- 1Department of Pathology, Soonchunhyang University Cheonan Hospital, Cheonan, Korea.
- 2Department of Pathology, Soonchunhyang University College of Medicine, Cheonan, Korea.
- 3Department of Surgery, Soonchunhyang University Cheonan Hospital, Cheonan, Korea. msslee@schmc.ac.kr
- KMID: 2372369
- DOI: http://doi.org/10.5230/jgc.2015.15.1.39
Abstract
- PURPOSE
Recent studies have revealed recurrent alterations in the cell adhesion gene FAT4, a candidate tumor suppressor gene, in cancer. FAT atypical cadherin 4 (FAT4) is a transmembrane receptor involved in the Hippo signaling pathway, which is involved in the control of organ size. Here, we investigated the loss of FAT4 expression and its association with clinicopathological risk factors in gastric cancer.
MATERIALS AND METHODS
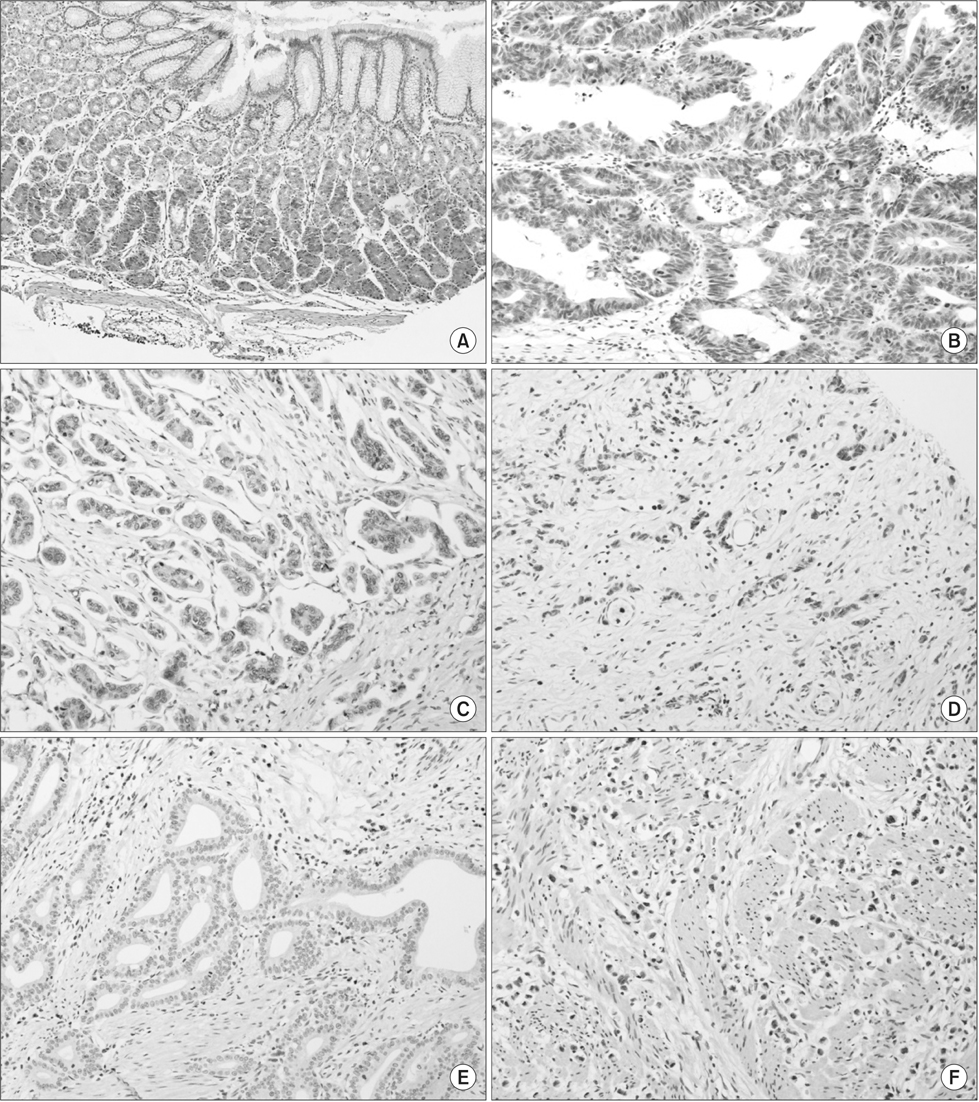
We assessed the expression of FAT4 by using immunohistochemistry on three tissue microarrays containing samples from 136 gastric cancer cases, radically resected in the Soonchunhyang University Cheonan Hospital between July 2006 and June 2008. Cytoplasmic immunoexpression of FAT4 was semi-quantitatively scored using the H-score system. An H-score of > or =10 was considered positive for FAT4 expression.
RESULTS
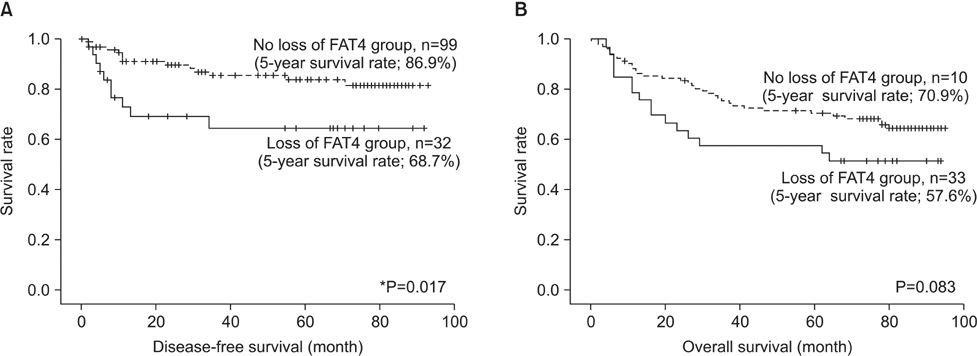
Variable cytoplasmic expressions of FAT4 were observed in gastric cancers, with 33 cases (24.3%) showing loss of expression (H-score <10). Loss of FAT4 expression was associated with an increased rate of perineural invasion (H-score <10 vs. > or =10, 36.4% vs. 16.5%, P=0.015), high pathologic T stage (P=0.015), high tumor-node-metastasis stage (P=0.017), and reduced disease-free survival time (H-score <10 vs. > or =10, mean survival 62.7+/-7.3 months vs. 79.1+/-3.1 months, P=0.025). However, no association was found between the loss of FAT4 expression and tumor size, gross type, histologic subtype, Lauren classification, lymphovascular invasion, or overall survival.
CONCLUSIONS
Loss of FAT4 expression appears to be associated with invasiveness in gastric cancer.
MeSH Terms
Figure
Reference
-
1. Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009; 472:467–477.2. Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012; 44:570–574.3. Bryant PJ, Huettner B, Held LI Jr, Ryerse J, Szidonya J. Mutations at the fat locus interfere with cell proliferation control and epithelial morphogenesis in Drosophila. Dev Biol. 1988; 129:541–554.4. Axelrod JD. Progress and challenges in understanding planar cell polarity signaling. Semin Cell Dev Biol. 2009; 20:964–971.5. Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006; 16:2090–2100.6. Katoh M. Function and cancer genomics of FAT family genes (review). Int J Oncol. 2012; 41:1913–1918.7. Conlon I, Raff M. Size control in animal development. Cell. 1999; 96:235–244.8. Johnston LA, Gallant P. Control of growth and organ size in Drosophila. Bioessays. 2002; 24:54–64.9. Valletta D, Czech B, Thasler WE, Müller M, Bosserhoff AK, Hellerbrand C. Expression and function of the atypical cadherin FAT1 in chronic liver disease. Biochem Biophys Res Commun. 2012; 426:404–408.10. Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006; 124:267–273.11. Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev. 2014; 34:190–221.12. Qi C, Zhu YT, Hu L, Zhu YJ. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer. 2009; 124:793–798.13. Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012; 33:287–296.14. Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010; 467:1114–1117.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prognostic Significance of E-cadherin Expression in Renal Cell Carcinoma
- Expression of E-cadherin according to the Presence of High Risk Prognostic Factors, Clinical Stages and Pathologic Types in Cervical Cancer Patients Treated by Radical Hysterectomy
- Role of Hepactocyte Growth Factor, Met, and E-cadherin in the Progression of Gastric Carcinomas
- Expression of E-cadherin and p53 Proteins in Gastric Adenocarcinoma
- Expression of E-cadherin and Catenin (alpha-, beta-catenin) in Endometrial Cancer and Atypical Complex Endometrial Hyperplasia