J Breast Cancer.
2014 Mar;17(1):1-7.
Patterns and Biologic Features of p53 Mutation Types in Korean Breast Cancer Patients
- Affiliations
-
- 1Department of Surgery, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea. gsjjoon@yuhs.ac
- 2Department of Laboratory Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea.
Abstract
- PURPOSE
The p53 gene is one of the most frequently mutated genes in breast cancer. We investigated the patterns and biologic features of p53 gene mutation and evaluated their clinical significance in Korean breast cancer patients.
METHODS
Patients who underwent p53 gene sequencing were included. Mutational analysis of exon 5 to exon 9 of the p53 gene was carried out using polymerase chain reaction-denaturing high performance liquid chromatography and direct sequencing.
RESULTS
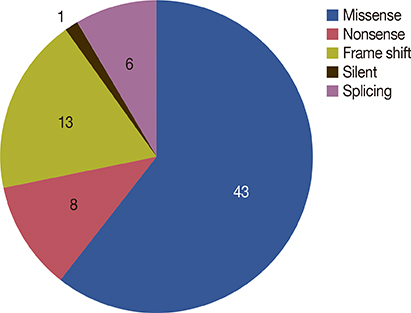
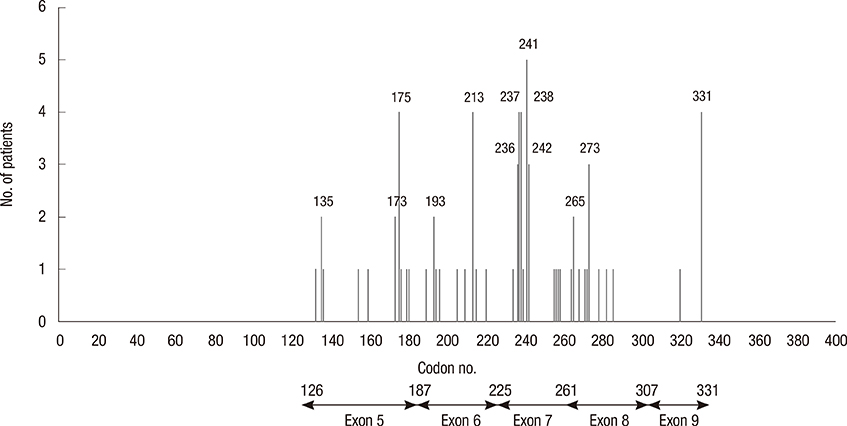
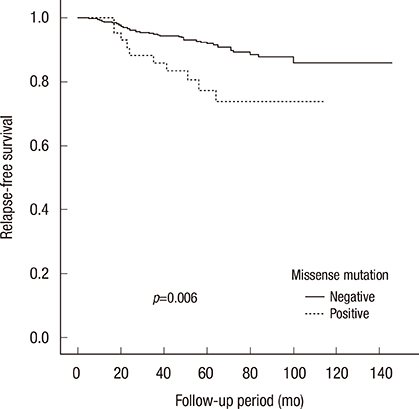
A total of 497 patients were eligible for the present study and p53 gene mutations were detected in 71 cases (14.3%). Mutation of p53 was significantly associated with histologic grading (p<0.001), estrogen receptor and progesterone receptor status (p<0.001), HER2 status (p<0.001), Ki-67 (p=0.028), and tumor size (p=0.004). The most frequent location of p53 mutations was exon 7 and missense mutation was the most common type of mutation. Compared with patients without mutation, there was a statistically significant difference in relapse-free survival of patients with p53 gene mutation and missense mutation (p=0.020, p=0.006, respectively). Only p53 missense mutation was an independent prognostic factor for relapse-free survival in multivariate analysis, with an adjusted hazard ratio of 2.29 (95% confidence interval, 1.08-4.89, p=0.031).
CONCLUSION
Mutation of the p53 gene was associated with more aggressive clinicopathologic characteristics and p53 missense mutation was an independent negative prognostic factor in Korean breast cancer patients.
Keyword
MeSH Terms
Figure
Reference
-
1. Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007; 109:25–32.
Article2. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, et al. Strategies for subtypes: dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011; 22:1736–1747.
Article3. Kaplan HG, Malmgren JA. Impact of triple negative phenotype on breast cancer prognosis. Breast J. 2008; 14:456–463.
Article4. Grann VR, Troxel AB, Zojwalla NJ, Jacobson JS, Hershman D, Neugut AI. Hormone receptor status and survival in a population-based cohort of patients with breast carcinoma. Cancer. 2005; 103:2241–2251.
Article5. Børresen-Dale AL. TP53 and breast cancer. Hum Mutat. 2003; 21:292–300.6. Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991; 351:453–456.
Article7. Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991; 352:345–347.
Article8. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.9. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000; 406:747–752.
Article10. Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003; 100:10393–10398.
Article11. Berns EM, van Staveren IL, Look MP, Smid M, Klijn JG, Foekens JA. Mutations in residues of TP53 that directly contact DNA predict poor outcome in human primary breast cancer. Br J Cancer. 1998; 77:1130–1136.
Article12. Olivier M, Langerød A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006; 12:1157–1167.
Article13. Pharoah PD, Day NE, Caldas C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer. 1999; 80:1968–1973.
Article14. Végran F, Rebucci M, Chevrier S, Cadouot M, Boidot R, Lizard-Nacol S. Only missense mutations affecting the DNA binding domain of p53 influence outcomes in patients with breast carcinoma. PLoS One. 2013; 8:e55103.
Article15. Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th ed. New York: Springer;2010.16. Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: a review. Hum Mutat. 2001; 17:439–474.
Article17. Keller G, Hartmann A, Mueller J, Höfler H. Denaturing high pressure liquid chromatography (DHPLC) for the analysis of somatic p53 mutations. Lab Invest. 2001; 81:1735–1737.
Article18. Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci. 2000; 113(Pt 10):1661–1670.
Article19. Olivier M, Hainaut P. TP53 mutation patterns in breast cancers: searching for clues of environmental carcinogenesis. Semin Cancer Biol. 2001; 11:353–360.
Article20. Berns EM, Foekens JA, Vossen R, Look MP, Devilee P, Henzen-Logmans SC, et al. Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res. 2000; 60:2155–2162.21. Bull SB, Ozcelik H, Pinnaduwage D, Blackstein ME, Sutherland DA, Pritchard KI, et al. The combination of p53 mutation and neu/erbB-2 amplification is associated with poor survival in node-negative breast cancer. J Clin Oncol. 2004; 22:86–96.
Article22. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012; 486:346–352.
Article23. Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res. 2000; 6:3923–3931.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation between p53 and MIB1 Index Expression of Primary Tumor and Metastatic Lymph Node in Breast Cancer
- Correlation between p53 and MIB1 Index Expression of Primary Tumor and Metastatic Lymph Node in Breast Cancer
- Sequencing of p53 Gene Mutations in Primary Breast Cancer Tissues
- A Case-control Study for Assessment of Risk Factors of Breast Cancer by the p53 Mutation
- Detecting p53 gene mutation of breast cancer and defining differences between silver staining PCR-SSCP and immunohistochemical staining