Nutr Res Pract.
2007 Mar;1(1):19-28.
Transcriptome analysis and promoter sequence studies on early adipogenesis in 3T3-L1 cells
- Affiliations
-
- 1Department of Biochemistry, College of Medicine, Hanyang University, Seoul 133-791, Korea.
- 2Department of Food Science and Human Nutrition, Research Institute of Human Ecology, Chonbuk National University, Jeonju, Jeonbuk 561-756, Korea. cha8@chonbuk.ac.kr
- 3Food Function Research, Division Korea Food Research Institute, San46-1, Baekhyun-dong, Bundang-gu, Sungnam-si, Gyeonggi-do 463-746, Korea.
Abstract
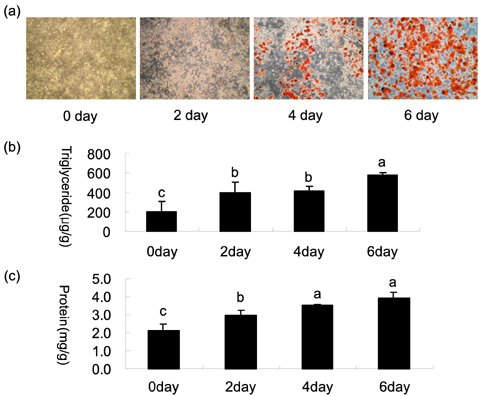
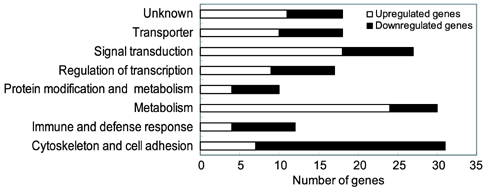
- To identify regulatory molecules which play key roles in the development of obesity, we investigated the transcriptional profiles in 3T3-L1 cells at early stage of differentiation and analyzed the promoter sequences of differentially regulated genes. One hundred and sixty-one (161) genes were found to have significant changes in expression at the 2nd day following treatment with differentiation cocktail. Among them, 86 transcripts were up-regulated and 75 transcripts were down-regulated. The 161 transcripts were classified into 10 categories according to their functional roles; cytoskeleton, cell adhesion, immune, defense response, metabolism, protein modification, protein metabolism, regulation of transcription, signal transduction and transporter. To identify transcription factors likely involved in regulating these differentially expressed genes, we analyzed the promoter sequences of up- or -down regulated genes for the presence of transcription factor binding sites (TFBSs). Based on coincidence of regulatory sites, we have identified candidate transcription factors (TFs), which include those previously known to be involved in adipogenesis (CREB, OCT-1 and c-Myc). Among them, c-Myc was also identified by our microarray data. Our approach to take advantage of the resource of the human genome sequences and the results from our microarray experiments should be validated by further studies of promoter occupancy and TF perturbation.
Keyword
MeSH Terms
Figure
Reference
-
1. Addya S, Keller MA, Delgrosso K, Ponte CM, Vadigepalli R, Gonye GE, Surrey S. Erythroid-induced commitment of K562 cells results in clusters of differentially expressed genes enriched for specific transcription regulatory elements. Physiol Genomics. 2004. 19:117–130.
Article2. Ahn JI, Lee KH, Shin DM, Shim JW, Lee JS, Chang SY, Lee YS, Brownstein MJ, Lee SH, Lee YS. Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons. Dev Biol. 2004. 265:491–501.
Article3. Berg AH, Lin Y, Lisanti MP, Scherer PE. Adipocyte differentiation induces dynamic changes in NF-kappaB expression and activity. Am J Physiol Endocrinol Metab. 2004. 287:E1178–E1188.4. Begley CG, Lipkowitz S, Gobel V, Mahon KA, Bertness V, Green AR, Gough NM, Kirsch IR. Molecular characterization of NSCL, a gene encoding a helix-loop-helix protein expressed in the developing nervous system. Proc Natl Acad Sci U S A. 1992. 89:38–42.
Article5. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976. 72:248–254.
Article6. Burton GR, Guan Y, Nagarajan R, McGehee RE Jr. Microarray analysis of gene expression during early adipocyte differentiation. Gene. 2002. 293:21–31.
Article7. Burton GR, Nagarajan R, Peterson CA, McGehee RE Jr. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004. 329:167–185.
Article8. Clamp M, Andrews D, Barker D, Bevan P, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Hubbard T, Kasprzyk A, Keefe D, Lehvaslaiho H, Iyer V, Melsopp C, Mongin E, Pettett R, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Birney E. Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res. 2003. 31:38–42.
Article9. Do MS, Nam SY, Hong SE, Kim KW, Duncan JS, Beattie JH, Trayhurn P. Metallothionein gene expression in human adipose tissue from lean and obese subjects. Horm Metab Res. 2002. 34:348–351.
Article10. Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998. 78:783–809.
Article11. Guo X, Liao K. Analysis of gene expression profile during 3T3-L1 preadipocyte differentiation. Gene. 2000. 251:45–53.
Article12. Heath VJ, Gillespie DA, Crouch DH. Inhibition of the terminal stages of adipocyte differentiation by cMyc. Exp Cell Res. 2000. 254:91–98.
Article13. Imagawa M, Tsuchiya T, Nishihara T. Identification of inducible genes at the early stage of adipocyte differentiation of 3T3-L1 cells. Biochem Biophys Res Commun. 1999. 254:299–305.
Article14. Ishida Y, Taniguchi H, Baba S. Rapid and transient induction of c-fos and c-myc during adipose conversion of 3T3-L1 cells: no relationship with 1 alpha,25-dihydroxyvitamine D3-suppressed adipogenesis. Kobe J Med Sci. 1988. 34:263–270.15. Kim JH, Kim HY, Lee YS. A novel method using edge detection for signal extraction from cDNA microarray image analysis. Exp Mol Med. 2001. 33:83–88.
Article16. Kim S, Sohn I, Ahn JI, Lee KH, Lee YS, Lee YS. Hepatic gene expression profiles in a long-term high-fat diet-induced obesity mouse model. Gene. 2004. 340:99–109.
Article17. Morin CL, Schlaepfer IR, Eckel RH. Tumor necrosis factor-alpha eliminates binding of NF-Y and an octamer-binding protein to the lipoprotein lipase promoter in 3T3-L1 adipocytes. J Clin Invest. 1995. 95:1684–1689.
Article18. Nishizuka M, Honda K, Tsuchiya T, Nishihara T, Imagawa M. RGS2 promotes adipocyte differentiation in the presence of ligand for peroxisome proliferator-activated receptor gamma. J Biol Chem. 2001. 276:29625–29627.
Article19. Ntambi JM, Young-Cheul K. Adipocyte differentiation and gene expression. J Nutr. 2000. 130:3122S–3126S.
Article20. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999. 284:143–147.
Article21. Pulverer B, Sommer A, McArthur GA, Eisenman RN, Luscher B. Analysis of Myc/Max/Mad network members in adipogenesis: inhibition of the proliferative burst and differentiation by ectopically expressed Mad1. J Cell Physiol. 2000. 183:399–410.
Article22. Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil Red O. Histochemistry. 1992. 97:493–497.
Article23. Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992. 6:439–453.
Article24. Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000. 16:145–171.
Article25. Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000. 14:1293–1307.
Article26. Schmidt C, Beyersmann D. Transient peaks in zinc and metallothionein levels during differentiation of 3T3-L1 cells. Arch Biochem Biophys. 1999. 364:91–98.
Article27. Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001. 276:34167–34174.28. Traythurn P, Duncan JS, Wood AM, Beattie JH. Regulation of metallothionein gene expression and secretion in rat adipocytes differentiated from preadipocytes in primary culture. Horm Metab Res. 2000a. 32:542–547.
Article29. Traythurn P, Duncan JS, Wood AM, Beattie JH. Metallothionein gene expression and secretion in white adipose tissue. Am J Physiol Regul Integr Comp Physiol. 2000b. 279:R2329–R2335.30. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001. 98:5116–5121.
Article31. Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001. 413:131–138.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- NF-kappaB is involved in the TNF-alpha induced inhibition of the differentiation of 3T3-L1 cells by reducing PPARg expression
- Pear pomace water extract inhibits adipogenesis and induces apoptosis in 3T3-L1 adipocytes
- Cryptotanshinone Inhibits Lipid Accumulation in Differentiating 3T3-L1 Preadipocytes by Down-regulating C/EBP-α, PPAR-γ, FAS, Perilipin A, and STAT-3
- Cryptotanshinone Inhibits Lipid Accumulation in Differentiating 3T3-L1 Preadipocytes by Down-regulating C/EBP-α, PPAR-γ, FAS, Perilipin A, and STAT-3
- Cryptotanshinone Inhibits Lipid Accumulation in Differentiating 3T3-L1 Preadipocytes by Down-regulating C/EBP-α, PPAR-γ, FAS, Perilipin A, and STAT-3