Healthc Inform Res.
2013 Mar;19(1):50-55. 10.4258/hir.2013.19.1.50.
AnsNGS: An Annotation System to Sequence Variations of Next Generation Sequencing Data for Disease-Related Phenotypes
- Affiliations
-
- 1Seoul National University Biomedical Informatics (SNUBI), Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea. juhan@snu.ac.kr
- 2Systems Biomedical Informatics Research Center, Seoul National University, Seoul, Korea.
- KMID: 2166663
- DOI: http://doi.org/10.4258/hir.2013.19.1.50
Abstract
OBJECTIVES
Next-generation sequencing (NGS) data in the identification of disease-causing genes provides a promising opportunity in the diagnosis of disease. Beyond the previous efforts for NGS data alignment, variant detection, and visualization, developing a comprehensive annotation system supported by multiple layers of disease phenotype-related databases is essential for deciphering the human genome. To satisfy the impending need to decipher the human genome, it is essential to develop a comprehensive annotation system supported by multiple layers of disease phenotype-related databases.
METHODS
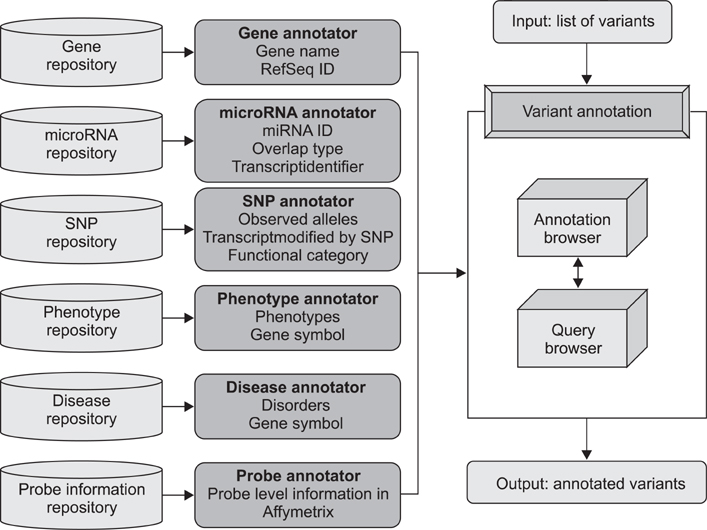
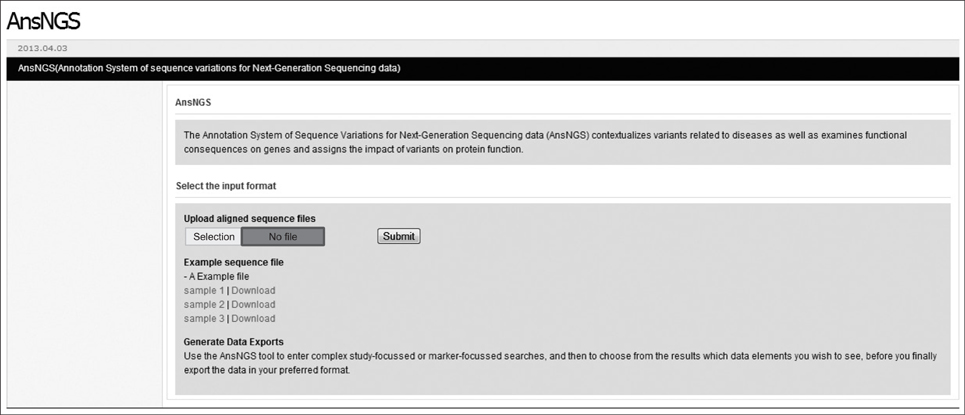
AnsNGS (Annotation system of sequence variations for next-generation sequencing data) is a tool for contextualizing variants related to diseases and examining their functional consequences. The AnsNGS integrates a variety of annotation databases to attain multiple levels of annotation.
RESULTS
The AnsNGS assigns biological functions to variants, and provides gene (or disease)-centric queries for finding disease-causing variants. The AnsNGS also connects those genes harbouring variants and the corresponding expression probes for downstream analysis using expression microarrays. Here, we demonstrate its ability to identify disease-related variants in the human genome.
CONCLUSIONS
The AnsNGS can give a key insight into which of these variants is already known to be involved in a disease-related phenotype or located in or near a known regulatory site. The AnsNGS is available free of charge to academic users and can be obtained from http://snubi.org/software/AnsNGS/.
Keyword
MeSH Terms
Figure
Reference
-
1. Kaiser J. DNA sequencing: a plan to capture human diversity in 1000 genomes. Science. 2008. 319(5862):395.2. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010. 38(16):e164.
Article3. Shetty AC, Athri P, Mondal K, Horner VL, Steinberg KM, Patel V, et al. SeqAnt: a Web service to rapidly identify and annotate DNA sequence variations. BMC Bioinformatics. 2010. 11:471.
Article4. Sana ME, Iascone M, Marchetti D, Palatini J, Galasso M, Volinia S. GAMES identifies and annotates mutations in next-generation sequencing projects. Bioinformatics. 2011. 27(1):9–13.
Article5. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002. 12(6):996–1006.
Article6. Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006. 342:129–138.
Article7. Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004. 36(5):431–432.
Article8. Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM). Hum Mutat. 2000. 15(1):57–61.
Article9. Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007. 4(11):907–909.
Article10. Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009. 461(7261):272–276.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- SFannotation: A Simple and Fast Protein Function Annotation System
- A Primer for Disease Gene Prioritization Using Next-Generation Sequencing Data
- A Simple GUI-based Sequencing Format Conversion Tool for the Three NGS Platforms
- CaGe: A Web-Based Cancer Gene Annotation System for Cancer Genomics
- Molecular genetic decoding of malformations of cortical development