Distribution of Malassezia Species on the Scalp in Korean Seborrheic Dermatitis Patients
- Affiliations
-
- 1Department of Dermatology, Konkuk University College of Medicine, Seoul, Korea.
- 2Department of Dermatology, Seoul National University College of Medicine, Seoul, Korea.
- 3Department of Dermatology, Chung-Ang University College of Medicine, Seoul, Korea. beomjoon@unitel.co.kr
- 4Department of Biological Sciences, Konkuk University, Seoul, Korea.
- 5College of Pharmacy, Chung-Ang University, Seoul, Korea.
- 6Department of Otorhinolaryngology-Head and Neck Surgery, Chung-Ang University College of Medicine, Seoul, Korea.
- 7Department of Emergency Medicine, Chung-Ang University College of Medicine, Seoul, Korea.
- 8Department of Genetic Engineering and Skin Biotechnology Center, Kyung Hee University, Seoul, Korea.
- KMID: 2156656
- DOI: http://doi.org/10.5021/ad.2011.23.2.156
Abstract
- BACKGROUND
Malassezia species play an important role in the pathogenesis of seborrheic dermatitis. In particular, M. restricta and M. globosa are considered to be the predominant organisms in seborrheic dermatitis of Western countries. However, species distribution of Malassezia in seborrheic dermatitis has not been clearly determined yet in Asia.
OBJECTIVE
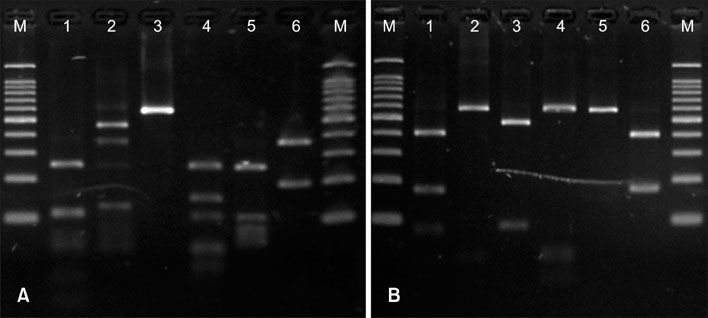
To identify the distribution of Malassezia species on the scalp of seborrheic dermatitis patients in Korea using 26S rDNA PCR-RFLP analysis.
METHODS
A total of 40 seborrheic dermatitis patients and 100 normal healthy volunteers were included in this study. For the identification of Malassezia species, the scalp scales of the subjects were analyzed by 26S rDNA PCR-RFLP analysis.
RESULTS
The most commonly identified Malassezia species were M. restricta in the seborrheic dermatitis patients, and M. globosa in the normal controls. In the seborrheic dermatitis group, M. restricta was identified in 47.5%, M. globosa in 27.5%, M. furfur in 7.5%, and M. sympodialis in 2.5% of patients. In the healthy control group, M. globosa was identified in 32.0%, M. restricta in 25.0%, M. furfur in 8.0%, M. obtusa in 6.0%, M. slooffiae in 6.0%, and M. sympodialis in 4.0% of subjects.
CONCLUSION
M. restricta is considered to be the most important Malassezia species in Korean seborrheic dermatitis patients.
Keyword
MeSH Terms
Figure
Cited by 4 articles
-
Evaluation of Expression of Lipases and Phospholipases of Malassezia restricta in Patients with Seborrheic Dermatitis
Yang Won Lee, Shin Yung Lee, Younghoon Lee, Won Hee Jung
Ann Dermatol. 2013;25(3):310-314. doi: 10.5021/ad.2013.25.3.310.Progress in Malassezia Research in Korea
Soo Young Kim, Yang Won Lee, Yong Beom Choe, Kyu Joong Ahn
Ann Dermatol. 2015;27(6):647-657. doi: 10.5021/ad.2015.27.6.647.Efficacy and Safety of Cream Containing Climbazole/Piroctone Olamine for Facial Seborrheic Dermatitis: A Single-Center, Open-Label Split-Face Clinical Study
Hae Jeong Youn, Soo Young Kim, Minji Park, Won Hee Jung, Yang Won Lee, Yong Beom Choe, Kyu Joong Ahn
Ann Dermatol. 2016;28(6):733-739. doi: 10.5021/ad.2016.28.6.733.In Vitro Anti-Malassezia Activity of Castanea crenata Shell and Oil-Soluble Glycyrrhiza Extracts
Song Hee Han, Min Seok Hur, Min Jung Kim, Won Hee Jung, Minji Park, Jeong Hwan Kim, Hong Ju Shin, Yong Beom Choe, Kyu Joong Ahn, Yang Won Lee
Ann Dermatol. 2017;29(3):321-326. doi: 10.5021/ad.2017.29.3.321.
Reference
-
1. Janik MP, Heffernan MP. Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. Yeast infections: candidiasis, pityriasis versicolor. Fitzpatrick's dermatology in general medicine. 2008. 7th ed. New York: McGraw-Hill;1822–1830.2. Guého E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996. 69:337–355.
Article3. Sugita T, Tajima M, Ito T, Saito M, Tsuboi R, Nishikawa A. Antifungal activities of tacrolimus and azole agents against the eleven currently accepted Malassezia species. J Clin Microbiol. 2005. 43:2824–2829.
Article4. Zisova LG. Malassezia species and seborrheic dermatitis. Folia Med (Plovdiv). 2009. 51:23–33.5. Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL Jr. Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004. 51:785–798.6. Morishita N, Sei Y, Sugita T. Molecular analysis of Malassezia microflora from patients with pityriasis versicolor. Mycopathologia. 2006. 161:61–65.
Article7. Sugita T, Tajima M, Tsubuku H, Tsuboi R, Nishikawa A. Quantitative analysis of cutaneous Malassezia in atopic dermatitis patients using real-time PCR. Microbiol Immunol. 2006. 50:549–552.
Article8. Gupta AK, Boekhout T, Theelen B, Summerbell R, Batra R. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J Clin Microbiol. 2004. 42:4253–4260.
Article9. Gandra RF, Simão RC, Matsumoto FE, da Silva BC, Ruiz LS, da Silva EG, et al. Genotyping by RAPD-PCR analyses of Malassezia furfur strains from pityriasis versicolor and seborrhoeic dermatitis patients. Mycopathologia. 2006. 162:273–280.
Article10. Guillot J, Deville M, Berthelemy M, Provost F, Guého E. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett Appl Microbiol. 2000. 31:400–403.
Article11. Makimura K, Tamura Y, Kudo M, Uchida K, Saito H, Yamaguchi H. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J Med Microbiol. 2000. 49:29–35.
Article12. Sugita T, Kodama M, Saito M, Ito T, Kato Y, Tsuboi R, et al. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J Clin Microbiol. 2003. 41:3022–3027.
Article13. Jang SJ, Lim SH, Ko JH, Oh BH, Kim SM, Song YC, et al. The investigation on the distribution of Malassezia yeasts on the normal Korean skin by 26S rDNA PCR-RFLP. Ann Dermatol. 2009. 21:18–26.
Article14. Lee YW, Kim SM, Oh BH, Lim SH, Choe YB, Ahn KJ. Isolation of 19 strains of Malassezia dermatis from healthy human skin in Korea. J Dermatol. 2008. 35:772–777.
Article15. González A, Sierra R, Cárdenas ME, Grajales A, Restrepo S, Cepero de Garcia MC, et al. Physiological and molecular characterization of atypical isolates of Malassezia furfur. J Clin Microbiol. 2009. 47:48–53.
Article16. Zomorodian K, Mirhendi H, Tarazooie B, Zeraati H, Hallaji Z, Balighi K. Distribution of Malassezia species in patients with psoriasis and healthy individuals in Tehran, Iran. J Cutan Pathol. 2008. 35:1027–1031.
Article17. Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005. 61:281–284.
Article18. Clift DC, Dodd HJ, Kirby JD, Midgley G, Noble WC. Seborrheic dermatitis and malignancy. An investigation of the skin flora. Acta Derm Venereol. 1988. 68:48–52.19. Lim SW, Shin MG, Lim JY, Yun SJ, Kim SJ, Lee SC, et al. Nested PCR for detection of Malassezia species from patient skin scales and clinical strains. Korean J Dermatol. 2008. 46:446–452.20. Sugita T, Suzuki M, Goto S, Nishikawa A, Hiruma M, Yamazaki T, et al. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med Mycol. 2010. 48:229–233.
Article21. Gaitanis G, Velegraki A, Alexopoulos EC, Chasapi V, Tsigonia A, Katsambas A. Distribution of Malassezia species in pityriasis versicolor and seborrhoeic dermatitis in Greece. Typing of the major pityriasis versicolor isolate M. globosa. Br J Dermatol. 2006. 154:854–859.
Article22. Tajima M, Sugita T, Nishikawa A, Tsuboi R. Molecular analysis of Malassezia microflora in seborrheic dermatitis patients: comparison with other diseases and healthy subjects. J Invest Dermatol. 2008. 128:345–351.
Article23. Takahata Y, Sugita T, Kato H, Nishikawa A, Hiruma M, Muto M. Cutaneous Malassezia flora in atopic dermatitis differs between adults and children. Br J Dermatol. 2007. 157:1178–1182.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Malassezia Species Cultured From the Lesions of Seborrheic Dermatitis
- Comparison of Clinical Manifestations and Distribution of Malassezia Species in Patients with Seborrheic Dermatitis and Atopic Dermatitis
- A Case of Seborrheic Dermatitis Caused by Overgrowth of Malassezia globosa
- Progress in Malassezia Research in Korea
- The Study on the Incidence of Pityriasis Versicolor of the Scalp in a Normal Person and Patients with Seborrheic Dermatitis