J Korean Med Sci.
2012 Oct;27(10):1129-1136. 10.3346/jkms.2012.27.10.1129.
Prediction of Microbial Infection of Cultured Cells Using DNA Microarray Gene-Expression Profiles of Host Responses
- Affiliations
-
- 1Seoul National University Biomedical Informatics (SNUBI), Seoul National University College of Medicine, Seoul, Korea. juhan@snu.ac.kr
- 2Department of Biochemistry and Molecular Biology, Seoul National University College of Medicine, Seoul, Korea.
- 3Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Korea.
- 4Biologics Headquater, Korea Food and Drug Administration, Seoul, Korea.
- 5Department of Microbiology College of Pharmacy, Chungbuk National University, Cheongju, Korea.
- 6Systems Biomedical Informatics National Core Research Center, Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 1778816
- DOI: http://doi.org/10.3346/jkms.2012.27.10.1129
Abstract
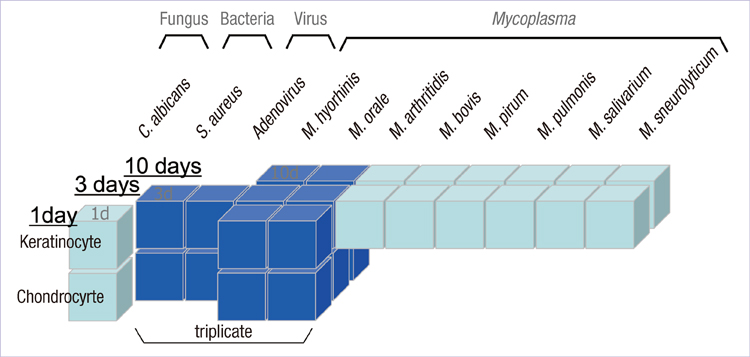
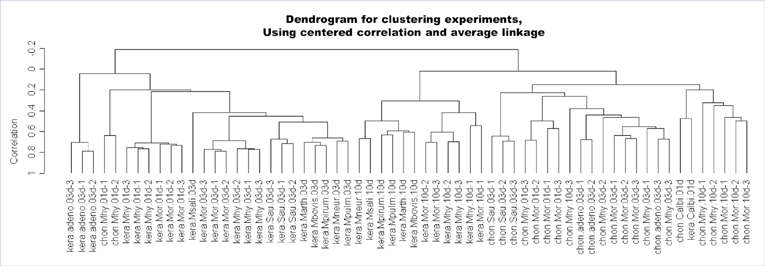
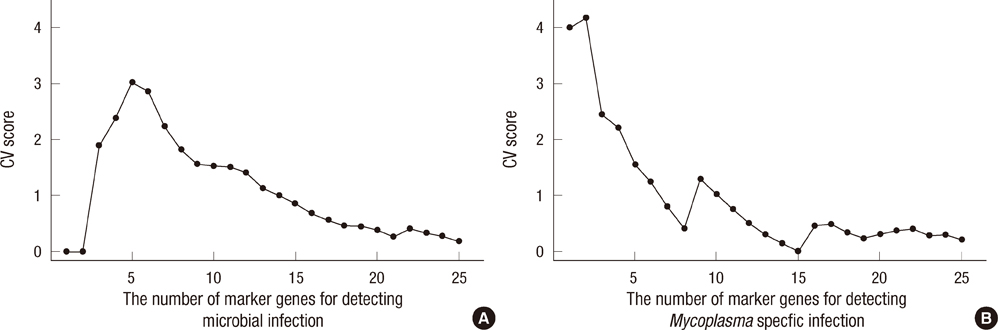
- Infection by microorganisms may cause fatally erroneous interpretations in the biologic researches based on cell culture. The contamination by microorganism in the cell culture is quite frequent (5% to 35%). However, current approaches to identify the presence of contamination have many limitations such as high cost of time and labor, and difficulty in interpreting the result. In this paper, we propose a model to predict cell infection, using a microarray technique which gives an overview of the whole genome profile. By analysis of 62 microarray expression profiles under various experimental conditions altering cell type, source of infection and collection time, we discovered 5 marker genes, NM_005298, NM_016408, NM_014588, S76389, and NM_001853. In addition, we discovered two of these genes, S76389, and NM_001853, are involved in a Mycolplasma-specific infection process. We also suggest models to predict the source of infection, cell type or time after infection. We implemented a web based prediction tool in microarray data, named Prediction of Microbial Infection (http://www.snubi.org/software/PMI).
MeSH Terms
Figure
Reference
-
1. Darin N, Kadhom N, Briere JJ, Chretien D, Bebear CM, Rotig A, Munnich A, Rustin P. Mitochondrial activities in human cultured skin fibroblasts contaminated by Mycoplasma hyorhinis. BMC Biochem. 2003. 4:15.2. Browning GF, Marenda MS, Noormohammadi AH, Markham PF. The central role of lipoproteins in the pathogenesis of mycoplasmoses. Vet Microbiol. 2011. 153:44–50.3. Rottem S. Interaction of mycoplasmas with host cells. Physiol Rev. 2003. 83:417–432.4. Cimolai N. Do mycoplasmas cause human cancer? Can J Microbiol. 2001. 47:691–697.5. Choi JW, Haigh WG, Lee SP. Caveat: mycoplasma arginine deiminase masquerading as nitric oxide synthase in cell cultures. Biochim Biophys Acta. 1998. 1404:314–320.6. Blanchard A, Bébéar CM. Shmuel R, Richard H, editors. Mycoplasmas of humans. Molecular biology and pathogenicity of mycoplasmas. 2002. New York, NY: Springer;45–71.7. Uuskula A, Kohl PK. Genital mycoplasmas, including Mycoplasma genitalium, as sexually transmitted agents. Int J STD AIDS. 2002. 13:79–85.8. Gilroy CB, Keat A, Taylor-Robinson D. The prevalence of Mycoplasma fermentans in patients with inflammatory arthritides. Rheumatology (Oxford). 2001. 40:1355–1358.9. Citti C, Nouvel LX, Baranowski E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 2010. 5:1073–1085.10. Mandruzzato S. Technological platforms for microarray gene expression profiling. Adv Exp Med Biol. 2007. 593:12–18.11. Park JY, Park YR, Park CH, Kim JH, Kim JH. Xperanto: a web-based integrated system for DNA microarray data management and analysis. Genomics Inform. 2005. 3:39–42.12. Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002. 18:Suppl 1. S96–S104.13. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002. 30:e15.14. Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol. 2001. 8:37–52.15. Stacey GN. Cell culture contamination. Methods Mol Biol. 2011. 731:79–91.16. Padua MB, Hansen PJ. Changes in expression of cell-cycle-related genes in PC-3 prostate cancer cells caused by ovine uterine serpin. J Cell Biochem. 2009. 107:1182–1188.17. Shi Z, Jervis D, Nickerson PE, Chow RL. Requirement for the paired-like homeodomain transcription factor VSX1 in type 3a mouse retinal bipolar cell terminal differentiation. J Comp Neurol. 2012. 520:117–129.18. Mizukami T, Kanai Y, Fujisawa M, Kanai-Azuma M, Kurohmaru M, Hayashi Y. Five azacytidine, a DNA methyltransferase inhibitor, specifically inhibits testicular cord formation and Sertoli cell differentiation in vitro. Mol Reprod Dev. 2008. 75:1002–1010.19. Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011. 378:1812–1823.20. Li Z, Zhang W, Wu M, Zhu S, Gao C, Sun L, Zhang R, Qiao N, Xue H, Hu Y, et al. Gene expression-based classification and regulatory networks of pediatric acute lymphoblastic leukemia. Blood. 2009. 114:4486–4493.21. Bhattacharjee M, Sillanpaa MJ. A bayesian mixed regression based prediction of quantitative traits from molecular marker and gene expression data. PLoS One. 2011. 6:e26959.22. van den Akker EB, Verbruggen B, Heijmans BT, Beekman M, Kok JN, Slagboom PE, Reinders MJ. Integrating protein-protein interaction networks with gene-gene co-expression networks improves gene signatures for classifying breast cancer metastasis. J Integr Bioinform. 2011. 8:188.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Detection of Mycoplasma Infection in Cultured Cells on the Basis of Molecular Profiling of Host Responses
- Differentially Expressed Cellular Gene Profiles between Healthy HIV-infected Koreans and AIDS Patients
- Gene Expression Analysis of Murine Primary Microglia Stimulated with LPS using Microarray
- Acetaminophen Induced Cytotoxicity and Altered Gene Expression in Cultured Cardiomyocytes of H9C2 Cells
- Basic Concept of Gene Microarray