J Bacteriol Virol.
2012 Jun;42(2):101-107. 10.4167/jbv.2012.42.2.101.
Our Genome and Our other Genome: Understanding humans as Symbionts with Microbes
- Affiliations
-
- 1The Laboratory of Human-Microbial Genomics, Department of Medicine, College of Medicine, Korea University, Anam-Dong, Seongbuk-Gu, Seoul, Korea. hstanleykim@korea.ac.kr
- KMID: 1717681
- DOI: http://doi.org/10.4167/jbv.2012.42.2.101
Abstract
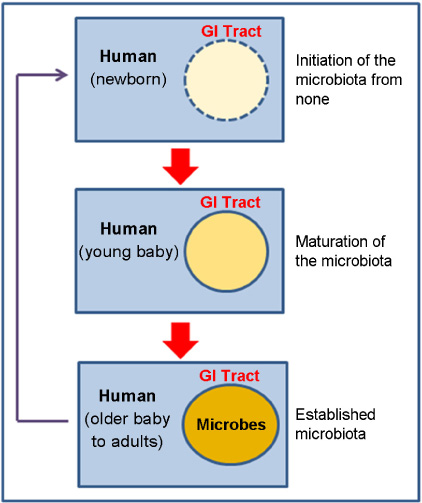
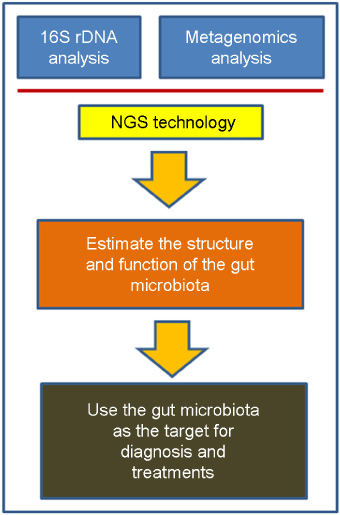
- One of the most significant discoveries in life sciences and medicine in recent years is that we humans are symbionts with a large number of microbes. These microbes reside on all over our surface, with the major portion of the population being in the digestive tract. The gut microbiota (microbial community) consists of up to 1,000 species of bacteria and its number exceeds ten-times of that of the human cells. Throughout the history of co-evolution, humans and microbes have become dependent on each other, and as the result developed a complex web of specific interactions. Recent development of the fast and cost-effective next generation sequencing (NGS) technology enabled the researchers to dissect the structure and function of the gut microbiota and their associations with human physiology in much detail. This newly-blooming field of human-microbial genomics will completely change the way we see and treat ourselves.
MeSH Terms
Figure
Cited by 1 articles
-
Roles of Enteric Microbial Composition and Metabolism in Health and Diseases
Jung Mogg Kim
Korean J Gastroenterol. 2013;62(4):191-205. doi: 10.4166/kjg.2013.62.4.191.
Reference
-
1. Mändar R, Mikelsaar M. Transmission of mother's microflora to the newborn at birth. Biol Neonate. 1996. 69:30–35.
Article2. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006. 118:511–521.
Article3. Grönlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999. 28:19–25.
Article4. Savage DC, Dubos R, Schaedler RW. Microbial ecology of hte gastrointestinal tract. Annu Rev Microbiol. 1977. 31:107–133.5. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004. 118:229–241.
Article6. Noverr MC, Huffinagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004. 12:562–568.
Article7. Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982. 15:189–203.
Article8. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007. 5:e177.
Article9. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006. 124:837–848.
Article10. Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006. 21:517–523.
Article11. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005. 307:1915–1920.
Article12. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005. 308:1635–1638.
Article13. Zhou J, Xia B, Treves DS, Wu LY, Marsh TL, O'Neill RV, et al. Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol. 2002. 68:326–334.
Article14. Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006. 59:1639–1650.
Article15. Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003. 185:6220–6223.
Article16. Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, David LA, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011. 480:241–244.
Article17. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011. 473:174–180.
Article18. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science. 2011. 334:105–108.
Article19. Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002. 22:283–307.
Article20. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003. 361:512–519.
Article21. Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci U S A. 2007. 104:10643–10648.
Article22. Human Microbiome Jumpstart Reference Strains Consortium. Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, et al. A Catalog of Reference Genomes from the Human Microbiome. Science. 2010. 328:994–999.23. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010. 464:59–65.
Article24. de Vos WM, Bron PA, Kleerebezem M. Post-genomics of lactic acid bacteria and other food-grade bacteria to discover gut functionality. Curr Opin Biotechnol. 2004. 15:86–93.
Article25. Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001. 294:2348–2351.
Article26. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004. 101:15718–15723.
Article27. McGarr SE, Ridlon JM, Hylemon PB. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol. 2005. 39:98–109.28. Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002. 35:S6–S16.
Article29. Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005. 307:1920–1925.
Article30. Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004. 53:685–693.
Article31. Hume G, Radford-Smith GL. The pathogenesis of Crohn's disease in the 21st century. Pathology. 2002. 34:561–567.32. Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003. 52:237–242.
Article33. Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A. 2004. 101:14919–14924.
Article34. Cerdeño-Tárraga AM, patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005. 307:1463–1465.
Article35. Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007. 5:e156.
Article36. Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003. 299:2074–2076.
Article37. Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006. 312:1355–1359.
Article38. Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001. 107:129–134.
Article39. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006. 444:1022–1023.40. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006. 444:1027–1031.
Article41. Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002. 68:219–226.
Article42. Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE, et al. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci U S A. 2002. 99:972–977.
Article43. Tannock GW, Munro K, Harmsen HJ, Welling GW, Smart J, Gopal PK. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. 2000. 66:2578–2588.
Article44. Kuehl CJ, Wood HD, Marsh TL, Schmidt TM, Young VB. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect Immun. 2005. 73:6952–6961.
Article