J Bacteriol Virol.
2012 Mar;42(1):49-55. 10.4167/jbv.2012.42.1.49.
Characterization of a Replication Element in the Coat Protein ORF of Turnip Yellow Mosaic Virus
- Affiliations
-
- 1Department of Biochemistry, Chungbuk National University, Cheongju, Korea. tjcho@chungbuk.ac.kr
- KMID: 1434772
- DOI: http://doi.org/10.4167/jbv.2012.42.1.49
Abstract
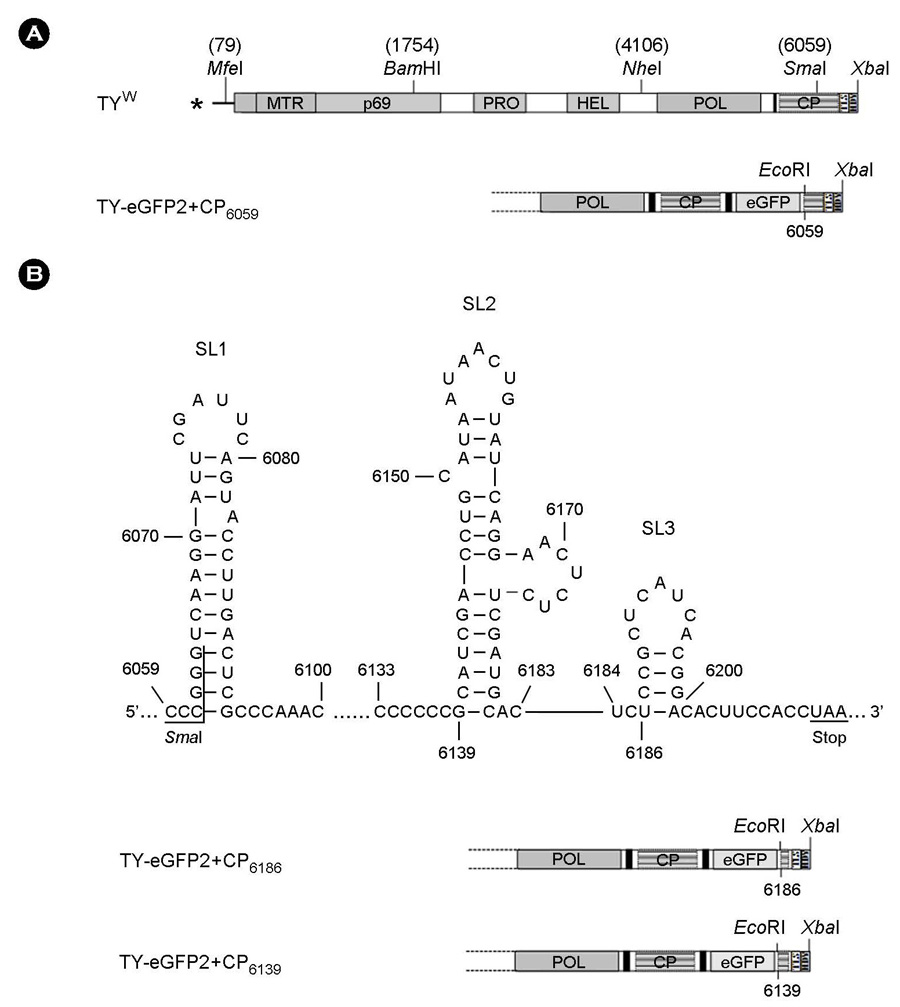
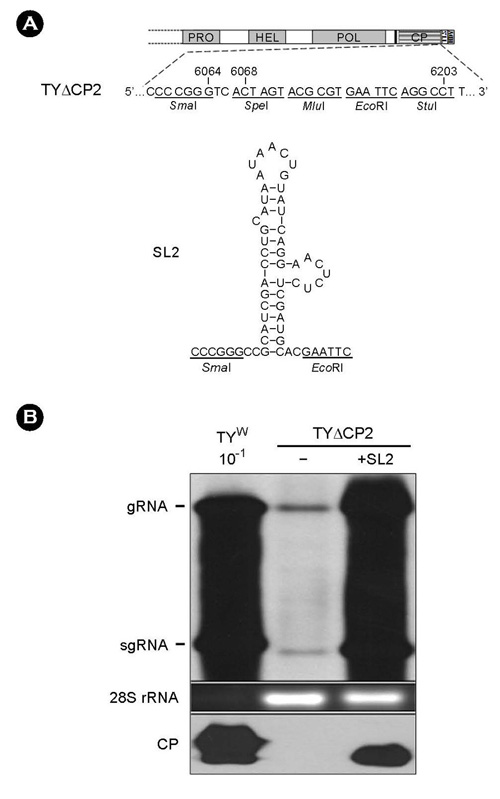
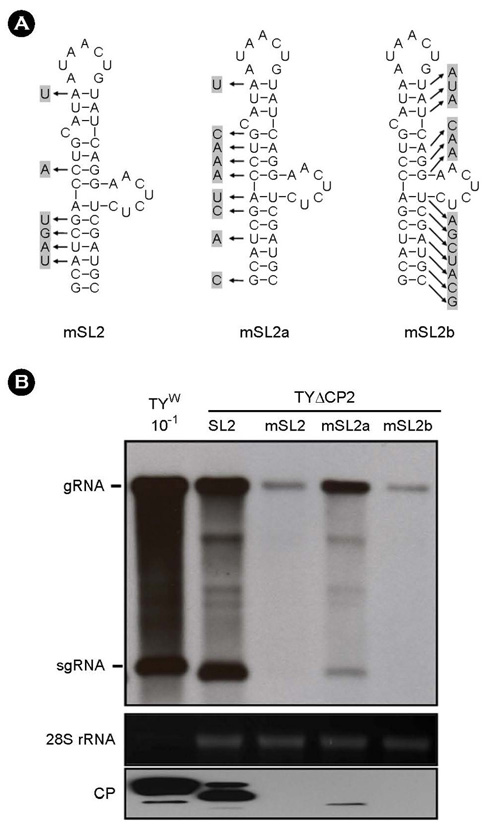
- Turnip yellow mosaic virus (TYMV) is a non-enveloped icosahedral virus that has a single 6.3 kb positive-strand RNA as a genome. Previously, it was observed that the recombinant construct TY-eGFP2, where an eGFP gene was inserted at the position downstream of the coat protein (CP) ORF of TYMV genome, barely replicated. The inhibition of replication was relieved by insertion of an additional copy of the 3' quarter of the CP ORF after the foreign sequence. In this study, we have examined if the 3' quarter of the CP ORF contains any replication elements. M-fold analysis predicted three stem-loop structures in this region. Analysis of the TY-eGFP2 constructs containing one or two of these stem-loop structures indicates that the secondary structure predicted in the region between nt-6139 and nt-6181, termed SL2, is essential for TYMV replication. The critical role of SL2 was confirmed by the observation that deletion of the 3' quarter of the CP ORF from the wild-type TYMV genome nearly abolished replication and that insertion of SL2 into the deletion mutant restored the replication. Mutations disrupting the stem of SL2 greatly reduced viral RNA replication, indicating that the secondary structure is essential for the enhancing activity.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Read-through Mutation in the Coat Protein ORF Suppresses Turnip Yellow Mosaic Virus Subgenomic RNA Accumulation
Hyun-Il Shin, Tae-Ju Cho
J Bacteriol Virol. 2013;43(1):54-63. doi: 10.4167/jbv.2013.43.1.54.
Reference
-
1. Dreher TW. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol. 2004. 5:367–375.
Article2. Matsuda D, Dreher TW. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3'-translational enhancer. Virology. 2004. 321:36–46.
Article3. Matsuda D, Bauer L, Tinnesand K, Dreher TW. Expression of the two nested overlapping reading frames of Turnip yellow mosaic virus RNA is enhanced by a 5' cap and by 5' and 3' viral sequences. J Virol. 2004. 78:9325–9335.
Article4. Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell. 2004. 16:1302–1313.
Article5. Singh RN, Dreher TW. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997. 233:430–439.
Article6. Deiman BA, Koenen AK, Verlaan PW, Pleij CW. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol. 1998. 72:3965–3972.
Article7. Deiman BA, Verlaan PW, Pleij CW. In vitro transcription by the turnip yellow mosaic virus RNA polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J Virol. 2000. 74:264–271.
Article8. Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCR- boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998. 4:1083–1095.
Article9. Hellendoorn K, Michiels PJ, Buitenhuis R, Pleij CW. Protonatable hairpins are conserved in the 5'-untranslated region of tymovirus RNAs. Nucleic Acids Res. 1996. 24:4910–4917.
Article10. Hellendoorn K, Verlaan PW, Pleij CW. A functional role for the conserved protonatable hairpins in the 5' untranslated region of turnip yellow mosaic virus RNA. J Virol. 1997. 71:8774–8779.
Article11. Shin HI, Cho NJ, Cho TJ. Role of 5'-UTR hairpins of the Turnip yellow mosaic virus RNA in replication and systemic movement. BMB Rep. 2008. 41:778–783.
Article12. Shin HI, Tzanetakis IE, Dreher TW, Cho TJ. The 5'-UTR of Turnip yellow mosaic virus does not include a critical encapsidation signal. Virology. 2009. 387:427–435.
Article13. Shin HI, Cho TJ. A sequence in coat protein open reading frame is required for Turnip yellow mosaic virus replication. J Bacteriol Virol. 2011. 41:109–116.
Article14. Cho TJ, Dreher TW. Encapsidation of genomic but not subgenomic turnip yellow mosaic virus RNA by coat protein provided in trans. Virology. 2006. 356:126–135.
Article15. Miller WA, White KA. Long-distance RNA-RNA interaction in plant virus gene expression and replication. Annu Rev Phytopathol. 2006. 44:447–467.
Article16. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003. 31:3406–3415.
Article17. Weiland JJ, Dreher TW. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc Natl Acad Sci U S A. 1993. 90:6095–6099.
Article18. Dreher TW. Functions of the 3'-untranslated regions of positive strand RNA viral genomes. Annu Rev Phytopathol. 1999. 37:151–174.
Article19. Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009. 1789:495–517.
Article20. Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Microbiol. 2006. 4:371–382.
Article21. Prod'homme D, Jakubiec A, Tournier V, Drugeon G, Jupin I. Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J Virol. 2003. 77:9124–9135.22. Jakubiec A, Notaise J, Tournier V, Héricourt F, Block MA, Drugeon G, et al. Assembly of turnip yellow mosaic virus replication complexes: Interaction between the proteinase and polymerase domains of the replication proteins. J Virol. 2004. 78:7945–7957.
Article23. Chen J, Noueiry A, Ahlquist P. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5' proximal RNA2 signal. J Virol. 2001. 75:3207–3219.
Article24. Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003. 41:77–98.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Sequence in Coat Protein Open Reading Frame Is Required for Turnip Yellow Mosaic Virus Replication
- Read-through Mutation in the Coat Protein ORF Suppresses Turnip Yellow Mosaic Virus Subgenomic RNA Accumulation
- Replication of Recombinant Flock House Virus RNA Encapsidated by Turnip Yellow Mosaic Virus Coat Proteins in Nicotiana benthamiana
- N-terminal Extension of Coat Protein of Turnip Yellow Mosaic Virus has Variable Effects on Replication, RNA Packaging, and Virion Assembly Depending on the Inserted Sequence
- Genome Size Constraint in Replication and Packaging of Turnip Yellow Mosaic Virus