Ann Lab Med.
2012 Sep;32(5):380-384. 10.3343/alm.2012.32.5.380.
A Case Report of Fanconi Anemia Diagnosed by Genetic Testing Followed by Prenatal Diagnosis
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea. MWSeong@snu.ac.kr
- 2Department of Pediatrics, Seoul National University Children's Hospital, Seoul, Korea.
- 3Department of Obstetrics and Gynecology, Seoul National University Hospital, Seoul, Korea.
- KMID: 1387356
- DOI: http://doi.org/10.3343/alm.2012.32.5.380
Abstract
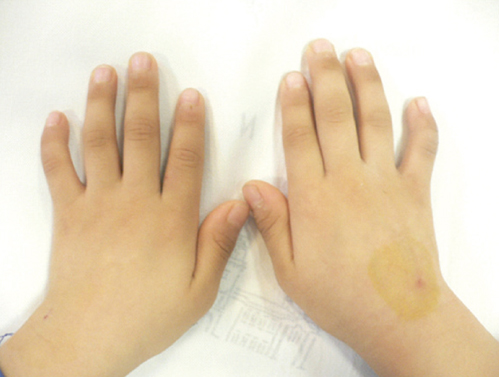
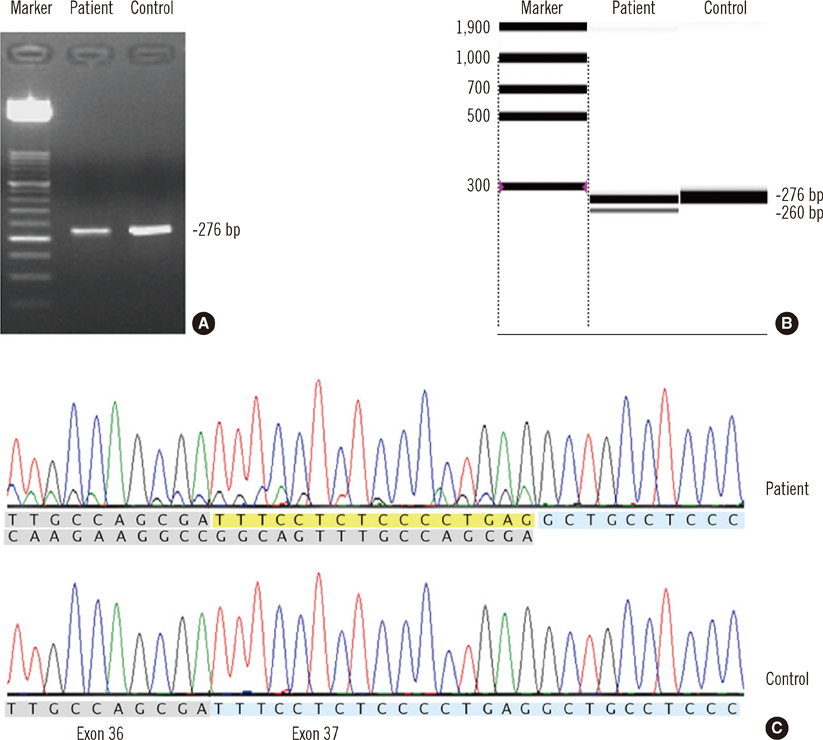
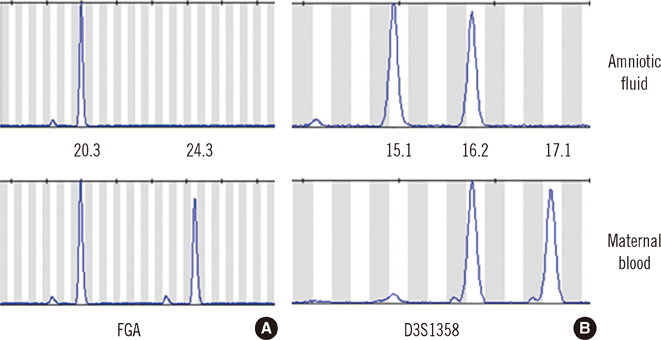
- Fanconi anemia (FA) is a rare genetic disorder affecting multiple body systems. Genetic testing, including prenatal testing, is a prerequisite for the diagnosis of many clinical conditions. However, genetic testing is complicated for FA because there are often many genes that are associated with its development, and large deletions, duplications, or sequence variations are frequently found in some of these genes. This study describes successful genetic testing for molecular diagnosis, and subsequent prenatal diagnosis, of FA in a patient and his family in Korea. We analyzed all exons and flanking regions of the FANCA, FANCC, and FANCG genes for mutation identification and subsequent prenatal diagnosis. Multiplex ligation-dependent probe amplification analysis was performed to detect large deletions or duplications in the FANCA gene. Molecular analysis revealed two mutations in the FANCA gene: a frameshift mutation c.2546delC and a novel splice-site mutation c.3627-1G>A. The FANCA mutations were separately inherited from each parent, c.2546delC was derived from the father, whereas c.3627-1G>A originated from the mother. The amniotic fluid cells were c.3627-1G>A heterozygotes, suggesting that the fetus was unaffected. This is the first report of genetic testing that was successfully applied to molecular diagnosis of a patient and subsequent prenatal diagnosis of FA in a family in Korea.
Keyword
MeSH Terms
-
Base Sequence
Child, Preschool
Exons
Fanconi Anemia/*diagnosis/genetics
Fanconi Anemia Complementation Group A Protein/genetics
Fanconi Anemia Complementation Group C Protein/genetics
Fanconi Anemia Complementation Group G Protein/genetics
Female
Frameshift Mutation
Genetic Testing
Heterozygote
Humans
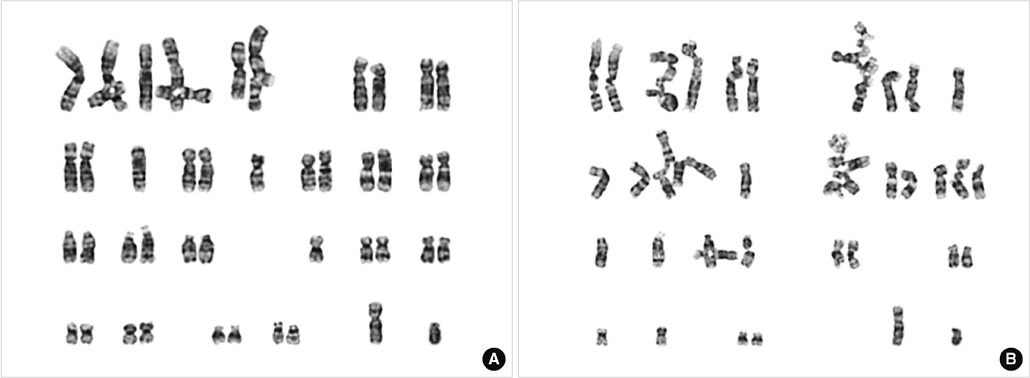
Karyotyping
Male
Pregnancy
Prenatal Diagnosis
RNA Splice Sites
Reverse Transcriptase Polymerase Chain Reaction
Sequence Analysis, DNA
Figure
Cited by 1 articles
-
Validation of Pathogenicity of Gene Variants in Fanconi Anemia Using Patient-derived Dermal Fibroblasts
Seon Young Park, Jong-Mi Lee, Myung-Jin Kim, Nack-Gyun Chung, Jung Bok Lee, Yonghwan Kim, Myungshin Kim
Ann Lab Med. 2023;43(1):127-131. doi: 10.3343/alm.2023.43.1.127.
Reference
-
1. Alter BP. Diagnosis, genetics, and management of inherited bone marrow failure syndromes. Hematology Am Soc Hematol Educ Program. 2007. 29–39.
Article2. Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006. 107:4223–4233.
Article3. Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood. 2003. 101:1249–1256.
Article4. Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003. 101:822–826.
Article5. Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010. 42:406–409.
Article6. Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011. 43:138–141.
Article7. Wijker M, Morgan NV, Herterich S, van Berkel CG, Tipping AJ, Gross HJ, et al. Heterogeneous spectrum of mutations in the Fanconi anaemia group A gene. Eur J Hum Genet. 1999. 7:52–59.
Article8. Bouchlaka C, Abdelhak S, Amouri A, Ben Abid H, Hadiji S, Frikha M, et al. Fanconi anemia in Tunisia: high prevalence of group A and identification of new FANCA mutations. J Hum Genet. 2003. 48:352–361.9. Callén E, Tischkowitz MD, Creus A, Marcos R, Bueren JA, Casado JA, et al. Quantitative PCR analysis reveals a high incidence of large intragenic deletions in the FANCA gene in Spanish Fanconi anemia patients. Cytogenet Genome Res. 2004. 104:341–345.
Article10. Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2005. 105:1329–1336.
Article11. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002. 30:e57.
Article12. Kook H, Cho D, Cho SH, Hong WP, Kim CJ, Park JY, et al. Fanconi anemia screening by diepoxybutane and mitomicin C tests in Korean children with bone marrow failure syndromes. J Korean Med Sci. 1998. 13:623–628.
Article13. Yamashita T, Nakahata T. Current knowledge on the pathophysiology of Fanconi anemia: from genes to phenotypes. Int J Hematol. 2001. 74:33–41.
Article14. Grompe M, D'Andrea A. Fanconi anemia and DNA repair. Hum Mol Genet. 2001. 10:2253–2259.
Article15. Tischkowitz MD, Hodgson SV. Fanconi anaemia. J Med Genet. 2003. 40:1–10.
Article16. Pinto FO, Leblanc T, Chamousset D, Le Roux G, Brethon B, Cassinat B, et al. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica. 2009. 94:487–495.
Article17. Bechtold A, Friedl R, Kalb R, Gottwald B, Neveling K, Gavvovidis I, et al. Prenatal exclusion/confirmation of Fanconi anemia via flow cytometry: a pilot study. Fetal Diagn Ther. 2006. 21:118–124.
Article