Korean J Radiol.
2011 Oct;12(5):588-594. 10.3348/kjr.2011.12.5.588.
Pre-Operative Prediction of Advanced Prostatic Cancer Using Clinical Decision Support Systems: Accuracy Comparison between Support Vector Machine and Artificial Neural Network
- Affiliations
-
- 1Department of Radiology, Seoul National University College of Medicine, Seoul 110-744, Korea. hakjlee@snu.ac.kr
- 2Department of Radiology, Seoul National University Hospital, Seoul 110-744, Korea.
- 3Department of Radiology, Research Institute and Hospital, National Cancer Center, Gyeonggi-do 410-769, Korea.
- 4Department of Radiology, Seoul National University Bundang Hospital, Gyeonggi-do 463-707, Korea.
- 5Department of Radiology, Seoul National University Boramae Hospital, Seoul 156-707, Korea.
- 6Department of Radiology, Kangwon National University College of Medicine, Gangwon-do 200-722, Korea.
- KMID: 1116444
- DOI: http://doi.org/10.3348/kjr.2011.12.5.588
Abstract
OBJECTIVE
The purpose of the current study was to develop support vector machine (SVM) and artificial neural network (ANN) models for the pre-operative prediction of advanced prostate cancer by using the parameters acquired from transrectal ultrasound (TRUS)-guided prostate biopsies, and to compare the accuracies between the two models.
MATERIALS AND METHODS
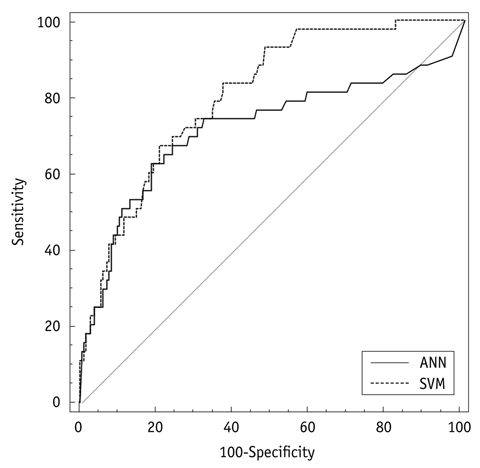
Five hundred thirty-two consecutive patients who underwent prostate biopsies and prostatectomies for prostate cancer were divided into the training and test groups (n = 300 versus n = 232). From the data in the training group, two clinical decision support systems (CDSSs-[SVM and ANN]) were constructed with input (age, prostate specific antigen level, digital rectal examination, and five biopsy parameters) and output data (the probability for advanced prostate cancer [> pT3a]). From the data of the test group, the accuracy of output data was evaluated. The areas under the receiver operating characteristic (ROC) curve (AUC) were calculated to summarize the overall performances, and a comparison of the ROC curves was performed (p < 0.05).
RESULTS
The AUC of SVM and ANN is 0.805 and 0.719, respectively (p = 0.020), in the pre-operative prediction of advanced prostate cancer.
CONCLUSION
The performance of SVM is superior to ANN in the pre-operative prediction of advanced prostate cancer.
Keyword
MeSH Terms
Figure
Reference
-
1. Chandana S, Leung H, Trpkov K. Staging of prostate cancer using automatic feature selection, sampling and Dempster-Shafer fusion. Cancer Inform. 2009. 7:57–73.2. Lee HJ, Hwang SI, Han SM, Park SH, Kim SH, Cho JY, et al. Image-based clinical decision support for transrectal ultrasound in the diagnosis of prostate cancer: comparison of multiple logistic regression, artificial neural network, and support vector machine. Eur Radiol. 2010. 20:1476–1484.3. Suzuki H, Komiya A, Kamiya N, Imamoto T, Kawamura K, Miura J, et al. Development of a nomogram to predict probability of positive initial prostate biopsy among Japanese patients. Urology. 2006. 67:131–136.4. Snow PB, Smith DS, Catalona WJ. Artificial neural networks in the diagnosis and prognosis of prostate cancer: a pilot study. J Urol. 1994. 152:1923–1926.5. Stephan C, Cammann H, Semjonow A, Diamandis EP, Wymenga LF, Lein M, et al. Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem. 2002. 48:1279–1287.6. Karakiewicz PI, Benayoun S, Kattan MW, Perrotte P, Valiquette L, Scardino PT, et al. Development and validation of a nomogram predicting the outcome of prostate biopsy based on patient age, digital rectal examination and serum prostate specific antigen. J Urol. 2005. 173:1930–1934.7. Chun FK, Briganti A, Graefen M, Montorsi F, Porter C, Scattoni V, et al. Development and external validation of an extended 10-core biopsy nomogram. Eur Urol. 2007. 52:436–444.8. Finne P, Finne R, Bangma C, Hugosson J, Hakama M, Auvinen A, et al. Algorithms based on prostate-specific antigen (PSA), free PSA, digital rectal examination and prostate volume reduce false-positive PSA results in prostate cancer screening. Int J Cancer. 2004. 111:310–315.9. Nam RK, Toi A, Klotz LH, Trachtenberg J, Jewett MA, Appu S, et al. Assessing individual risk for prostate cancer. J Clin Oncol. 2007. 25:3582–3588.10. Bianco FJ Jr. Nomograms and medicine. Eur Urol. 2006. 50:884–886.11. Loch T, Leuschner I, Genberg C, Weichert-Jacobsen K, Kuppers F, Yfantis E, et al. Artificial neural network analysis (ANNA) of prostatic transrectal ultrasound. Prostate. 1999. 39:198–204.12. Cortes C, Vapnik V. Support vector networks. Mach Learn. 1995. 20:273–297.13. Park EA, Lee HJ, Kim KG, Kim SH, Lee SE, Choe GY. Prediction of pathological stages before prostatectomy in prostate cancer patients: analysis of 12 systematic prostate needle biopsy specimens. Int J Urol. 2007. 14:704–708.14. Jiang L, Manry MT. Nonlinear networks for classification. Accessed on Aug 12, 2011. ftp.simtel.net/pub/simtelnet/msdos/calculte/Nuclass706a.zip.15. Comak E, Arslan A, Turkoglu I. A decision support system based on support vector machines for diagnosis of the heart valve diseases. Comput Biol Med. 2007. 37:21–27.16. Chang C-C, Lin C-J. LIBSVM-A library for support vector machines. Accessed on May 22, 2010. http://www.csie.ntu.edu.tw/~cjlin/libsvm.17. McNeal JE. Cancer volume and site of origin of adenocarcinoma in the prostate: relationship to local and distant spread. Hum Pathol. 1992. 23:258–266.18. Gancarczyk KJ, Wu H, McLeod DG, Kane C, Kusuda L, Lance R, et al. Using the percentage of biopsy cores positive for cancer, pretreatment PSA, and highest biopsy Gleason sum to predict pathologic stage after radical prostatectomy: the Center for Prostate Disease Research nomograms. Urology. 2003. 61:589–595.19. Sebo TJ, Bock BJ, Cheville JC, Lohse C, Wollan P, Zincke H. The percent of cores positive for cancer in prostate needle biopsy specimens is strongly predictive of tumor stage and volume at radical prostatectomy. J Urol. 2000. 163:174–178.20. Wills ML, Sauvageot J, Partin AW, Gurganus R, Epstein JI. Ability of sextant biopsies to predict radical prostatectomy stage. Urology. 1998. 51:759–764.21. Gohji K, Okamoto M, Takenaka A, Nomi M, Fujii A. Predicting the extent of prostate cancer using the combination of systematic biopsy and serum prostate-specific antigen in Japanese men. BJU Int. 1999. 83:39–42.22. Errejon A, Crawford ED, Dayhoff J, O'Donnell C, Tewari A, Finkelstein J, et al. Use of artificial neural networks in prostate cancer. Mol Urol. 2001. 5:153–158.23. Zlotta AR, Remzi M, Snow PB, Schulman CC, Marberger M, Djavan B. An artificial neural network for prostate cancer staging when serum prostate specific antigen is 10 ng./ml. or less. J Urol. 2003. 169:1724–1728.24. Babaian RJ, Fritsche H, Ayala A, Bhadkamkar V, Johnston DA, Naccarato W, et al. Performance of a neural network in detecting prostate cancer in the prostate-specific antigen reflex range of 2.5 to 4.0 ng/mL. Urology. 2000. 56:1000–1006.25. Vapnik V. Statistical learning theory, Wiley series on adaptive and learning systems for signal processing, communications and control. 1998. New York: John Wiley & Sons.26. Huang YL, Chen DR. Support vector machines in sonography: application to decision making in the diagnosis of breast cancer. Clin Imaging. 2005. 29:179–184.27. Moradi M, Abolmaesumi P, Siemens DR, Sauerbrei EE, Boag AH, Mousavi P. Augmenting detection of prostate cancer in transrectal ultrasound images using SVM and RF time series. IEEE Trans Biomed Eng. 2009. 56:2214–2224.28. Zhu Y, Tan Y, Hua Y, Wang M, Zhang G, Zhang J. Feature selection and performance evaluation of support vector machine (SVM)-based classifier for differentiating benign and malignant pulmonary nodules by computed tomography. J Digit Imaging. 2010. 23:51–65.29. Pochet NL, Suykens JA. Support vector machines versus logistic regression: improving prospective performance in clinical decision-making. Ultrasound Obstet Gynecol. 2006. 27:607–608.30. Byvatov E, Fechner U, Sadowski J, Schneider G. Comparison of support vector machine and artificial neural network systems for drug/nondrug classification. J Chem Inf Comput Sci. 2003. 43:1882–1889.31. Chang RF, Wu WJ, Moon WK, Chou YH, Chen DR. Support vector machines for diagnosis of breast tumors on US images. Acad Radiol. 2003. 10:189–197.32. Ravery V, Schmid HP, Toublanc M, Boccon-Gibod L. Is the percentage of cancer in biopsy cores predictive of extracapsular disease in T1-T2 prostate carcinoma? Cancer. 1996. 78:1079–1084.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Machine Learning on Early Diagnosis of Depression
- Anesthesia research in the artificial intelligence era
- Comparison of Accuracies for Image-based 1.5T and 3T MRI Using a Clinical Decision Support System Driven by a Support Vector Machine to Detect Seminal Vesicle Invasion of Prostate Cance
- A Comparison of Intensive Care Unit Mortality Prediction Models through the Use of Data Mining Techniques
- Gastrointestinal polyp detection in endoscopic images using an improved feature extraction method