Korean J Lab Med.
2011 Jan;31(1):37-43. 10.3343/kjlm.2011.31.1.37.
Application of Single-nucleotide Polymorphism and Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats Analyses to Clinical Mycobacterium tuberculosis Isolates from Korea
- Affiliations
-
- 1Department of Laboratory Medicine, Pusan National University, Busan, Korea.
- 2Department of Laboratory Medicine, Konyang University Hospital, College of Medical Science, Konyang University, Daejeon, Korea.
- 3Department of Thoracic and Cardiovascular Surgery, School of Medicine, Pusan National University, Busan, Korea.
- 4Department of Dermatology, School of Medicine, Pusan National University, Busan, Korea. drkmp@hanmail.net
- 5Medical Research Institute, Pusan National University, Busan, Korea.
- KMID: 1096658
- DOI: http://doi.org/10.3343/kjlm.2011.31.1.37
Abstract
- BACKGROUND
Single-nucleotide polymorphism (SNP) analysis is a powerful strategy for large-scale molecular population studies examining phylogenetic relationships among bacterial strains. Mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) can be easily digitized to share data among laboratories. This study applied SNP and MIRU-VNTR analyses for molecular strain typing of Mycobacterium tuberculosis isolates collected throughout Korea.
METHODS
We studied 102 clinical M. tuberculosis isolates, including 6 paired strains, collected from 11 university hospitals in Korea in 2008 and 2009. SNPs were detected using hairpin primer assays, and then, MIRU-VNTR analysis was performed.
RESULTS
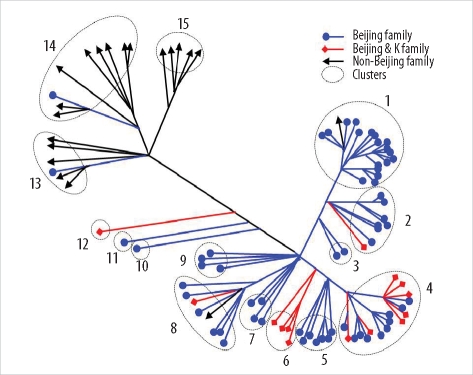
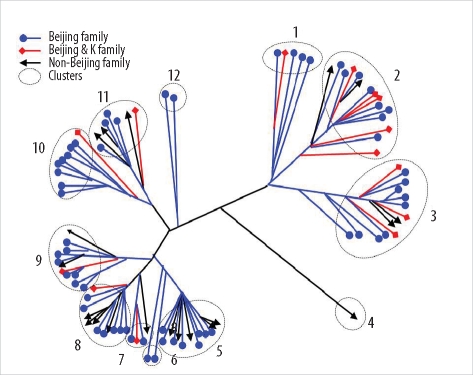
Thirty-five SNPs contained polymorphisms that helped differentiate the 96 tested isolates. The isolates were classified into 15 clusters. The Beijing family strains were distributed within closely related clusters in the SNP dendrogram. For MIRU-VNTR analysis, the 96 isolates were divided into 12 groups. The discriminatory index in 8 of these groups (MIRU-10, -23, -26, and -31; ETR-A, -B, -C, and -F) was high (Hunter-Gaston diversity index > 0.6). Unlike the SNP method, MIRU-VNTR analysis did not identify any notable localizations of Beijing or non-Beijing family isolates in specific clusters.
CONCLUSIONS
SNP and MIRU-VNTR analyses are surrogate molecular strain-typing methods for M. tuberculosis in Korea where Beijing family isolates are predominant.
Keyword
MeSH Terms
Figure
Reference
-
1. Gopaul KK, Brown TJ, Gibson AL, Yates MD, Drobniewski FA. Progression toward an improved DNA amplification-based typing technique in the study of Mycobacterium tuberculosis epidemiology. J Clin Microbiol. 2006; 44:2492–2498. PMID: 16825370.2. Van Soolingen D. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J Intern Med. 2001; 249:1–26. PMID: 11168781.
Article3. Seidler A, Nienhaus A, Diel R. The transmission of tuberculosis in the light of new molecular biological approaches. Occup Environ Med. 2004; 61:96–102. PMID: 14739374.
Article4. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993; 31:406–409. PMID: 8381814.5. Barlow RE, Gascoyne-Binzi DM, Gillespie SH, Dickens A, Qamer S, Hawkey PM. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J Clin Microbiol. 2001; 39:2453–2457. PMID: 11427553.6. Gori A, Bandera A, Marchetti G, Degli Esposti A, Catozzi L, Nardi GP, et al. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis. 2005; 11:1242–1248. PMID: 16102314.7. Gori A, Esposti AD, Bandera A, Mezzetti M, Sola C, Marchetti G, et al. Comparison between spoligotyping and IS6110 restriction fragment length polymorphisms in molecular genotyping analysis of Mycobacterium tuberculosis strains. Mol Cell Probes. 2005; 19:236–244. PMID: 16038791.8. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997; 35:907–914. PMID: 9157152.9. Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PW, Martin C, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999; 37:2607–2618. PMID: 10405410.10. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 393:537–544. PMID: 9634230.11. Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002; 184:5479–5490. PMID: 12218036.12. Kersulyte D, Mukhopadhyay AK, Velapatino B, Su W, Pan Z, Garcia C, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000; 182:3210–3218. PMID: 10809702.13. Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet. 2000; 58:250–264. PMID: 11076050.
Article14. Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, Cousins DV, et al. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics. 2002; 162:1533–1543. PMID: 12524330.15. Gutacker MM, Mathema B, Soini H, Shashkina E, Kreiswirth BN, Graviss EA, et al. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis. 2006; 193:121–128. PMID: 16323140.16. Frothingham R, Meeker-O'Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998; 144(Pt 5):1189–1196. PMID: 9611793.17. Choi GE, Jang MH, Song EJ, Jeong SH, Kim JS, Lee WG, et al. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for investigating epidemiologic distribution in Korea. J Korean Med Sci. 2010; 25:1716–1721. PMID: 21165284.18. Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbón MH, Bobadilla del Valle M, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006; 188:759–772. PMID: 16385065.19. Hazbón MH, Alland D. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J Clin Microbiol. 2004; 42:1236–1242. PMID: 15004082.20. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006; 44:4498–4510. PMID: 17005759.21. Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000; 36:762–771. PMID: 10844663.22. Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988; 26:2465–2466. PMID: 3069867.
Article23. Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, et al. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003; 3:125–133. PMID: 12809807.24. Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci U S A. 2004; 101:4871–4876. PMID: 15041743.25. Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002; 99:3684–3689. PMID: 11891304.26. Kaboev OK, Luchkina LA, Tret'iakov AN, Bahrmand AR. PCR hot start using primers with the structure of molecular beacons (hairpin-like structure). Nucleic Acids Res. 2000; 28:E94. PMID: 11058144.
Article27. Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 2002; 30:e37. PMID: 11972352.
Article28. Bouakaze C, Keyser C, de Martino SJ, Sougakoff W, Veziris N, Dabernat H, et al. Identification and genotyping of Mycobacterium tuberculosis complex species by use of a SNaPshot Minisequencing-based assay. J Clin Microbiol. 2010; 48:1758–1766. PMID: 20220173.29. Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001; 39:3563–3571. PMID: 11574573.30. Kwara A, Schiro R, Cowan LS, Hyslop NE, Wiser MF, Roahen Harrison S, et al. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003; 41:2683–2685. PMID: 12791904.31. Yun KW, Song EJ, Choi GE, Hwang IK, Lee EY, Chang CL. Strain typing of Mycobacterium tuberculosis isolates from Korea by mycobacterial interspersed repetitive units-variable number of tandem repeats. Korean J Lab Med. 2009; 29:314–319. PMID: 19726893.32. Hanekom M, van der Spuy GD, Gey van Pittius NC, McEvoy CR, Hoek KG, Ndabambi SL, et al. Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of Mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J Clin Microbiol. 2008; 46:3338–3345. PMID: 18716230.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Strain Typing of Mycobacterium tuberculosis Isolates from Korea by Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats
- Single-nucleotide polymorphismbased epidemiological analysis of Korean Mycobacterium bovis isolates
- Evaluation of the Selected 12-locus MIRU for Genotyping Beijing Family Mycobacterium tuberculosis in Korea
- Molecular Strain Typing of Mycobacterium tuberculosis: a Review of Frequently Used Methods
- The Present and Future of Molecular Epidemiology in Tuberculosis