J Vet Sci.
2014 Sep;15(3):381-388. 10.4142/jvs.2014.15.3.381.
Protective efficacy of a high-growth reassortant swine H3N2 inactivated vaccine constructed by reverse genetic manipulation
- Affiliations
-
- 1Division of Swine Infectious Diseases, Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai 200241, China. haiyu@shvri.ac.cn
- KMID: 2155621
- DOI: http://doi.org/10.4142/jvs.2014.15.3.381
Abstract
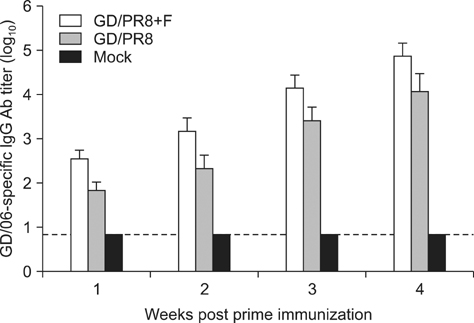
- Novel reassortant H3N2 swine influenza viruses (SwIV) with the matrix gene from the 2009 H1N1 pandemic virus have been isolated in many countries as well as during outbreaks in multiple states in the United States, indicating that H3N2 SwIV might be a potential threat to public health. Since southern China is the world's largest producer of pigs, efficient vaccines should be developed to prevent pigs from acquiring H3N2 subtype SwIV infections, and thus limit the possibility of SwIV infection at agricultural fairs. In this study, a high-growth reassortant virus (GD/PR8) was generated by plasmid-based reverse genetics and tested as a candidate inactivated vaccine. The protective efficacy of this vaccine was evaluated in mice by challenging them with another H3N2 SwIV isolate [A/Swine/Heilongjiang/1/05 (H3N2) (HLJ/05)]. Prime and booster inoculation with GD/PR8 vaccine yielded high-titer serum hemagglutination inhibiting antibodies and IgG antibodies. Complete protection of mice against H3N2 SwIV was observed, with significantly reduced lung lesion and viral loads in vaccine-inoculated mice relative to mock-vaccinated controls. These results suggest that the GD/PR8 vaccine may serve as a promising candidate for rapid intervention of H3N2 SwIV outbreaks in China.
MeSH Terms
-
Animals
Female
Influenza A Virus, H3N2 Subtype/*genetics/immunology
Influenza Vaccines/genetics/immunology/*therapeutic use
Mice
Mice, Inbred BALB C
Orthomyxoviridae Infections/immunology/*prevention & control/virology
Reassortant Viruses/genetics/immunology
Reverse Genetics/methods/*veterinary
Swine
Swine Diseases/immunology/*prevention & control/virology
Vaccines, Inactivated
Virus Replication
Influenza Vaccines
Vaccines, Inactivated
Figure
Cited by 1 articles
-
A novel M2e-multiple antigenic peptide providing heterologous protection in mice
Feng Wen, Ji-Hong Ma, Hai Yu, Fu-Ru Yang, Meng Huang, Yan-Jun Zhou, Ze-Jun Li, Xiu-Hui Wang, Guo-Xin Li, Yi-Feng Jiang, Wu Tong, Guang-Zhi Tong
J Vet Sci. 2016;17(1):71-78. doi: 10.4142/jvs.2016.17.1.71.
Reference
-
1. Booy R, Khandaker G, Heron LG, Yin J, Doyle B, Tudo KK, Hueston L, Gilbert GL, Macintyre CR, Dwyer DE. Cross-reacting antibodies against the pandemic (H1N1) 2009 influenza virus in older Australians. Med J Aust. 2011; 194:19–23.
Article2. Centers for Disease Control and Prevention (CDC). Swine-origin influenza A (H3N2) virus infection in two children--Indiana and Pennsylvania, July-August 2011. MMWR Morb Mortal Wkly Rep. 2011; 60:1213–1215.3. Centers for Disease Control and Prevention (CDC). Update: Influenza A (H3N2)v transmission and guidelines - five states, 2011. MMWR Morb Mortal Wkly Rep. 2012; 60:1741–1744.4. Chen H, Subbarao K, Swayne D, Chen Q, Lu X, Katz J, Cox N, Matsuoka Y. Generation and evaluation of a high-growth reassortant H9N2 influenza A virus as a pandemic vaccine candidate. Vaccine. 2003; 21:1974–1979.
Article5. Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, García-Sastre A, Palese P, Hai R. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol. 2011; 85:11235–11241.
Article6. Fan X, Zhu H, Zhou B, Smith DK, Chen X, Lam TT, Poon LL, Peiris M, Guan Y. Emergence and dissemination of a swine H3N2 reassortant influenza virus with 2009 pandemic H1N1 genes in pigs in China. J Virol. 2012; 86:2375–2378.
Article7. Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, García-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999; 73:9679–9682.
Article8. Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009; 325:197–201.
Article9. Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009; 361:1945–1952.
Article10. Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine. 2002; 20:3165–3170.
Article11. Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD, Bystrom JM, Alexandersen S, Pasick JM, Berhane Y, Morrison ME, Keenliside JM, Laurendeau S, Rohonczy EB. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J. 2009; 50:1153–1161.12. Karasin AI, Schutten MM, Cooper LA, Smith CB, Subbarao K, Anderson GA, Carman S, Olsen CW. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000; 68:71–85.
Article13. Lakdawala SS, Lamirande EW, Suguitan AL Jr, Wang W, Santos CP, Vogel L, Matsuoka Y, Lindsley WG, Jin H, Subbarao K. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog. 2011; 7:e1002443.
Article14. Lin SC, Huang MH, Tsou PC, Huang LM, Chong P, Wu SC. Recombinant trimeric HA protein immunogenicity of H5N1 avian influenza viruses and their combined use with inactivated or adenovirus vaccines. PLoS One. 2011; 6:e20052.
Article15. Lina B, Bouscambert M, Enouf V, Rousset D, Valette M, van der Werf S. S-OtrH3N2 viruses: use of sequence data for description of the molecular characteristics of the viruses and their relatedness to previously circulating H3N2 human viruses. Euro Surveill. 2011; 16:20039.
Article16. Liu Q, Ma J, Liu H, Qi W, Anderson J, Henry SC, Hesse RA, Richt JA, Ma W. Emergence of novel reassortant H3N2 swine influenza viruses with the 2009 pandemic H1N1 genes in the United States. Arch Virol. 2012; 157:555–562.
Article17. Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999; 73:5903–5911.
Article18. Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999; 96:9345–9350.
Article19. Pearce MB, Jayaraman A, Pappas C, Belser JA, Zeng H, Gustin KM, Maines TR, Sun X, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc Natl Acad Sci U S A. 2012; 109:3944–3949.
Article20. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938; 27:493–497.21. Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 "Spanish" influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A. 1999; 96:1651–1656.
Article22. Shortridge KF, Stuart-Harris CH. An influenza epicentre? Lancet. 1982; 2:812–813.
Article23. Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977; 196:1454–1455.
Article24. Skowronski DM, De Serres G, Janjua NZ, Gardy JL, Gilca V, Dionne M, Hamelin ME, Rhéaume C, Boivin G. Cross-reactive antibody to swine influenza A(H3N2) subtype virus in children and adults before and after immunisation with 2010/11 trivalent inactivated influenza vaccine in Canada, August to November 2010. Euro Surveill. 2012; 17:pii20066.
Article25. Skowronski DM, Janjua NZ, De Serres G, Purych D, Gilca V, Scheifele DW, Dionne M, Sabaiduc S, Gardy JL, Li G, Bastien N, Petric M, Boivin G, Li Y. Cross-reactive and vaccine-induced antibody to an emerging swine-origin variant of influenza A virus subtype H3N2 (H3N2v). J Infect Dis. 2012; 206:1852–1861.
Article26. Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009; 459:1122–1125.
Article27. Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA. 2004; 292:1333–1340.
Article28. Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003; 289:179–186.
Article29. Tian G, Zhang S, Li Y, Bu Z, Liu P, Zhou J, Li C, Shi J, Yu K, Chen H. Protective efficacy in chickens, geese and ducks of an H5N1-inactivated vaccine developed by reverse genetics. Virology. 2005; 341:153–162.
Article30. Tremblay D, Allard V, Doyon JF, Bellehumeur C, Spearman JG, Harel J, Gagnon CA. Emergence of a new swine H3N2 and pandemic (H1N1) 2009 influenza A virus reassortant in two Canadian animal populations, mink and swine. J Clin Microbiol. 2011; 49:4386–4390.
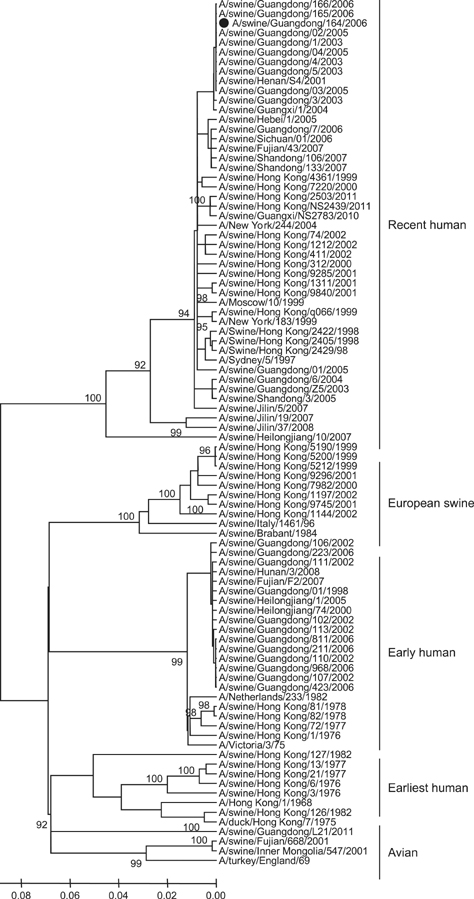
Article31. Yu H, Hua RH, Zhang Q, Liu TQ, Liu HL, Li GX, Tong GZ. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol. 2008; 46:1067–1075.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A novel M2e-multiple antigenic peptide providing heterologous protection in mice
- Reverse genetic platform for inactivated and live-attenuated influenza vaccine
- Requirements for improved vaccines against foot-and-mouth disease epidemics
- Inactivated genotype 1 Japanese encephalitis vaccine for swine
- Genetic identification and serological evaluation of commercial inactivated foot-and-mouth disease virus vaccine in pigs