Korean J Lab Med.
2008 Apr;28(2):95-102. 10.3343/kjlm.2008.28.2.95.
Detection of Rifampin Resistant Mycobacterium tuberculosis complex using Denaturing HPLC
- Affiliations
-
- 1Departmnt of Chemistry, School of Advanced Science and Basic Science Research Institute Dankook University, Cheonan, Korea.
- 2GeNet Bio Chungnam Animal Science Center, Konyang University, Nonsan, Korea.
- 3Department of Laboratory Medicine, Dankook University Hospital, Cheonan, Korea.
- 4Departmnt of Laboratory Medicine, Dankook University College of Medicine, Cheonan, Korea. wan1818@paran.com
- KMID: 854835
- DOI: http://doi.org/10.3343/kjlm.2008.28.2.95
Abstract
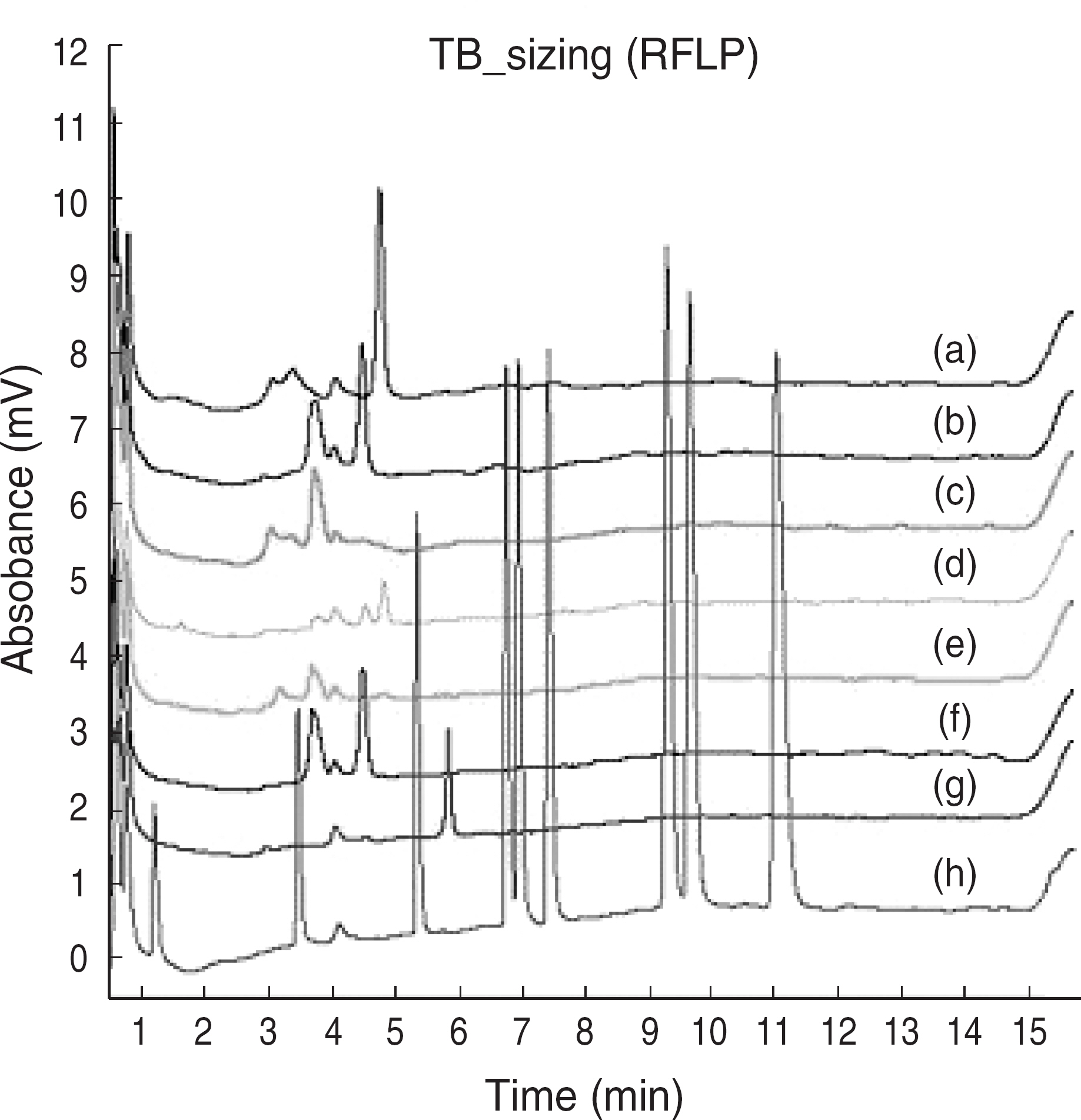
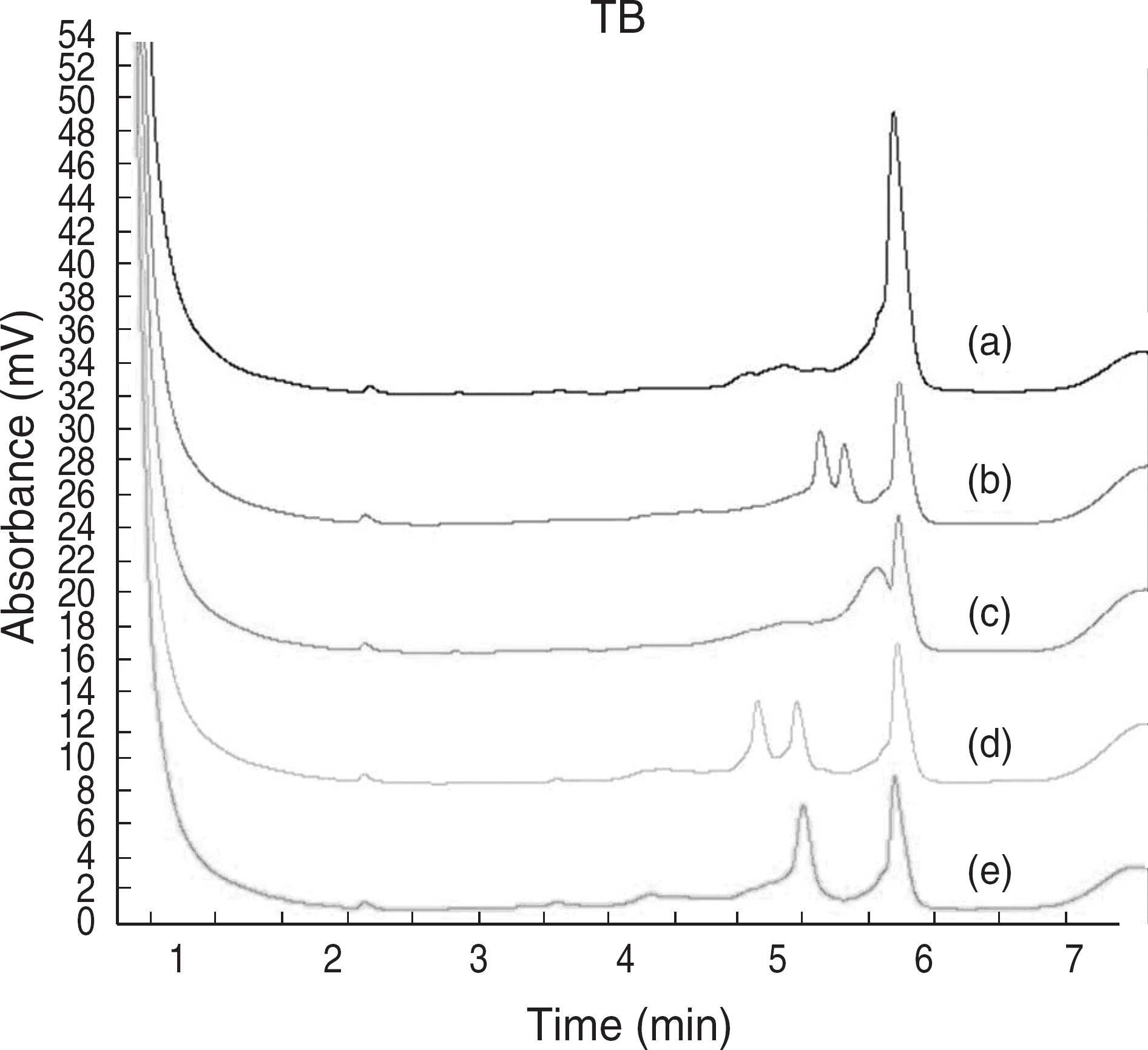
- BACKGROUND: Tuberculosis (TB) remains an important cause of morbidity and mortality throughout the world. The surge of TB has been accompanied by an increase in multi-drug-resistant tuberculosis (MDR-TB). In this study, we developed a denaturing HPLC (DHPLC) method for detecting rpoB gene mutation as a rifampin resistance based on sequence. METHODS: In this study, we used 99 mycobacterial isolates grown in Ogawa media. At first, we used a PCR method that can amplify the 235 bp and 136 bp rpoB DNAs of Mycobacterium tuberculosis complex (MTB) and Non-tuberculous mycobacteria (NTM). And then, PCR-restriction fragment length polymorphism (RFLP) of rpoB DNA (342 bp), which comprises the Rif(T) region, was used for the differential identification of Mycobacteria. Finally, we detected these amplicons by DHPLC, compared to PCR-RFLP results, and performed sequencing. RESULTS: Among 99 mycobacterial isolates, 80 (81%) were MTB and 19 (19%) were NTM. NTM were identified to 7 different species by DHPLC and PCR-RFLP. rpoB mutation was detected in 9 (11%) of the MTB specimens. These results were confirmed by using sequencing. CONCLUSIONS: DHPLC provided a rapid, simple, and automatable performance for detection of rifampin resistant Mycobacterium tuberculosis complex and would be helpful as a supplemental method in high-throughput clinical laboratories.
Keyword
MeSH Terms
Figure
Reference
-
1.Nunn P., Reid A., De Cock KM. Tuberculosis and HIV infection: the global setting. J Infect Dis. 2007. 196(S):S5–14.
Article2.Korea National Statistical Office. Annual report on the cause of death statistics (Based on vital registration). 2003. (통계청. 사망원인통계연보. 2003.).3.Barnes PF., Bloch AB., Davidson PT., Snider DE Jr. Tuberculosis in patients with human immuno-deficiency virus infection. N Engl J Med. 1991. 324:1644–50.
Article4.Cantwell MF., Snider DE Jr., Cauthen GM., Onorato IM. Epidemiology of tuberculosis in the United States, 1985 through 1992. JAMA. 1994. 272:535–9.
Article5.Starke JR., Jacobs RF., Jereb J. Resurgence of tuberculosis in children. J Pediatr. 1992. 120:839–55.
Article6.Morris S., Bai GH., Suffys P., Portillo-Gomez L., Fairchok M., Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995. 171:954–60.
Article7.Espinal MA., Laszlo A., Simonsen L., Boulahbal F., Kim SJ., Reniero A, et al. Global trends in resistance to antituberculosis drugs. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 2001. 344:1294–303.8.Hunt JM., Roberts GD., Stockman L., Felmlee TA., Persing DH. Detection of a genetic locus encoding resistance to rifampin in mycobacterial cultures and in clinical specimens. Diagn Microbiol Infect Dis. 1994. 18:219–27.
Article9.Donnabella V., Martiniuk F., Kinney D., Bacerdo M., Bonk S., Hanna B, et al. Isolation of the gene for the beta subunit of RNA polymerase from rifampicin-resistant Mycobacterium tuberculosis and identification of new mutations. Am J Respir Cell Mol Biol. 1994. 11:639–43.
Article10.Goble M., Iseman MD., Madsen LA., Waite D., Ackerson L., Horsburgh CR Jr. Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med. 1993. 328:527–32.
Article11.Lee MK., Park AJ., Jeon HS. Rapid Detection of rifampin Resistant Mycobacterium tuberculosis using the Lind probe Assay. Korean J Clin Pathol. 1997. 17:269–78. (이미경, 박애자, 전희선. Line probe assay를이용한 rifampin 내성 결핵균의 조기검출. 대한임상병리학회지 1997;17: 269-78.).12.Underhill PA., Jin L., Zemans R., Oefner PJ., Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA. 1996. 93:196–200.
Article13.Nam YH., Park SB., Lee JS., Kang W., Kim JG. The development of analysis system for genes related disease using chemical properties of DHPLC. Korean J Chem Soc. 2006. 50:116–22. (남윤형, 박상범, 이재식, 강원, 김종규. DHPLC의 화학적 특성을 이용한 질병 유전자의 분석시스템개발. 대한화학회지 2006;50: 116-22.).14.Kim BJ., Lee KH., Park BN., Kim SJ., Bai GH., Kim SJ, et al. Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB). J Clin Microbiol. 2001. 39:2102–9.15.Kim BJ., Hong SK., Lee KH., Yun YJ., Kim EC., Park YG, et al. Differential identification of Mycobacterium tuberculosis complex and non-tuberculous mycobacteria by duplex PCR assay using the RNA polymerase gene (rpoB). J Clin Microbiol. 2004. 42:1308–12.16.Kam KM., Yip CW. Surveillance of Mycobacterium tuberculosis drug resistance in Hong Kong, 1986-1999, after the implementation of directly observed treatment. Int J Tuberc Lung Dis. 2001. 5:815–23.17.Xiao W., Oefner PJ. Denaturing high-performance liquid chromatography: A review. Hum Mutat. 2001. 17:439–74.
Article18.Skopek TR., Glaab WE., Monroe JJ., Kort KL., Schaefer W. Analysis of sequence alterations in a defined DNA region: comparison of temperature-modulated heteroduplex analysis and denaturing gradient gel electrophoresis. Mutat Res. 1999. 430:13–21.
Article19.Jones AC., Austin J., Hansen N., Hoogendoorn B., Oefner PJ., Cheadle JP, et al. Optimal temperature selection for mutation detection by denaturing HPLC and comparison to single-stranded conformation polymorphism and heteroduplex analysis. Clin Chem. 1999. 45:1133–40.
Article20.Yip CW., Leung KL., Wong D., Cheung DT., Chu MY., Tang HS, et al. Denaturing HPLC for high-throughput screening of rifampicin-resistant Mycobacterium tuberculosis isolates. Int J Tuberc Lung Dis. 2006. 10:625–30.21.Wagner T., Stoppa-Lyonnet D., Fleischmann E., Muhr D., Pages S., Sandberg T, et al. Denaturing high-performance liquid chromatography detects reliably BRCA1 and BRCA2 mutations. Genomics. 1999. 62:369–76.
Article22.Ellis LA., Taylor CF., Taylor GR. A comparison of fluorescent SSCP and denaturing HPLC for high throughput mutation scanning. Hum Mutat. 2000. 15:556–64.
Article23.Gross E., Arnold N., Pfeifer K., Bandick K., Kiechle M. Identification of specific BRCA1 and BRCA2 variants by DHPLC. Hum Mutat. 2000. 16:345–53.24.Nickerson ML., Weirich G., Zbar B., Schmidt LS. Signature-based analysis of MET proto-oncogene mutations using DHPLC. Hum Mutat. 2000. 16:68–76.
Article25.Benit P., Kara-Mostefa A., Berthelon M., Sengmany K., Munnich A., Bonnefont JP. Mutation analysis of the hamartin gene using denaturing high performance liquid chromatography. Hum Mutat. 2000. 16:417–21.26.Lee JY., Choi HJ., Lee HY., Joung EY., Huh JW., Oh YM, et al. Recovery rate and characteristics of nontuberculous mycobacterial isolates in a university hospital in Korea. Tuberc Respir Dis. 2005. 58:385–91. (이정연, 최희진, 이혜영, 정은영, 허진원, 오연목 등. 한 대학병원에서 비결핵항산균의분리및동정실태. 결핵및호흡기질환 2005;58: 385-91.).
Article27.Yip CW., Leung KL., Wong D., Cheung DT., Chu MY., Tang HS, et al. Denaturing HPLC for high-throughput screening of rifampicin-resistant Mycobacterium tuberculosis isolates. Int J Tuberc Lung Dis. 2006. 10:625–30.28.Telenti A., Imboden P., Marchesi F., Lowrie D., Cole S., Colston MJ, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993. 341:647–50.29.Suh JT., Kim MH., Choi JW., Lee HJ., Kang HM. Direct sequencing for rpoB gene in rifampin-resistant Mycobacterium tuberculosis. Korean J Clin Pathol. 1996. 16:78–90. (서진태, 김문희, 최종원, 이희주, 강홍모. Mycobacterium tuberculosis의 rifampin 내성과관련된 rpoB 유전자염기서열분석. 대한임상병리학회지 1996;16: 78-90.).30.Lee MA. Direct detection of rifampin-resistant mycobacterium tuberculosis by polymerase chain reaction and line probe assay in sputums. Korean J Clin Pathol. 1998. 18:71–6. (이미애. 객담에서 중합효소연쇄반응과 Line Probe Assay를 이용한 Rifampin 내성결핵균의 직접검출법. 대한임상병리학회지 1998;18: 71-6.).
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Detection of Rifampin-Resistant Mycobacterium tuberculosis by Polymerase Chain Reaction and Single - Strand Conformation Polymorphism Analysis
- Use of RNA/RNA Duplex, Base Pair-mismatch Assay and 3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl Tetrazolium Bromide (MTT) Assay for Rapid Detection of Rifampin-resistant Mycobacterium tuberculosis
- Detection of Rifampin-resistance in Mycobacterium tuberculosis
- Rifampin-resistant Relapsed Tuberculosis Confirmed by Molecular Technique
- Rapid Detection of Rifampin Resistant Mycobacterium tuberculosis Using the Line Probe Assay