Ann Lab Med.
2024 Nov;44(6):621-624. 10.3343/alm.2024.0221.
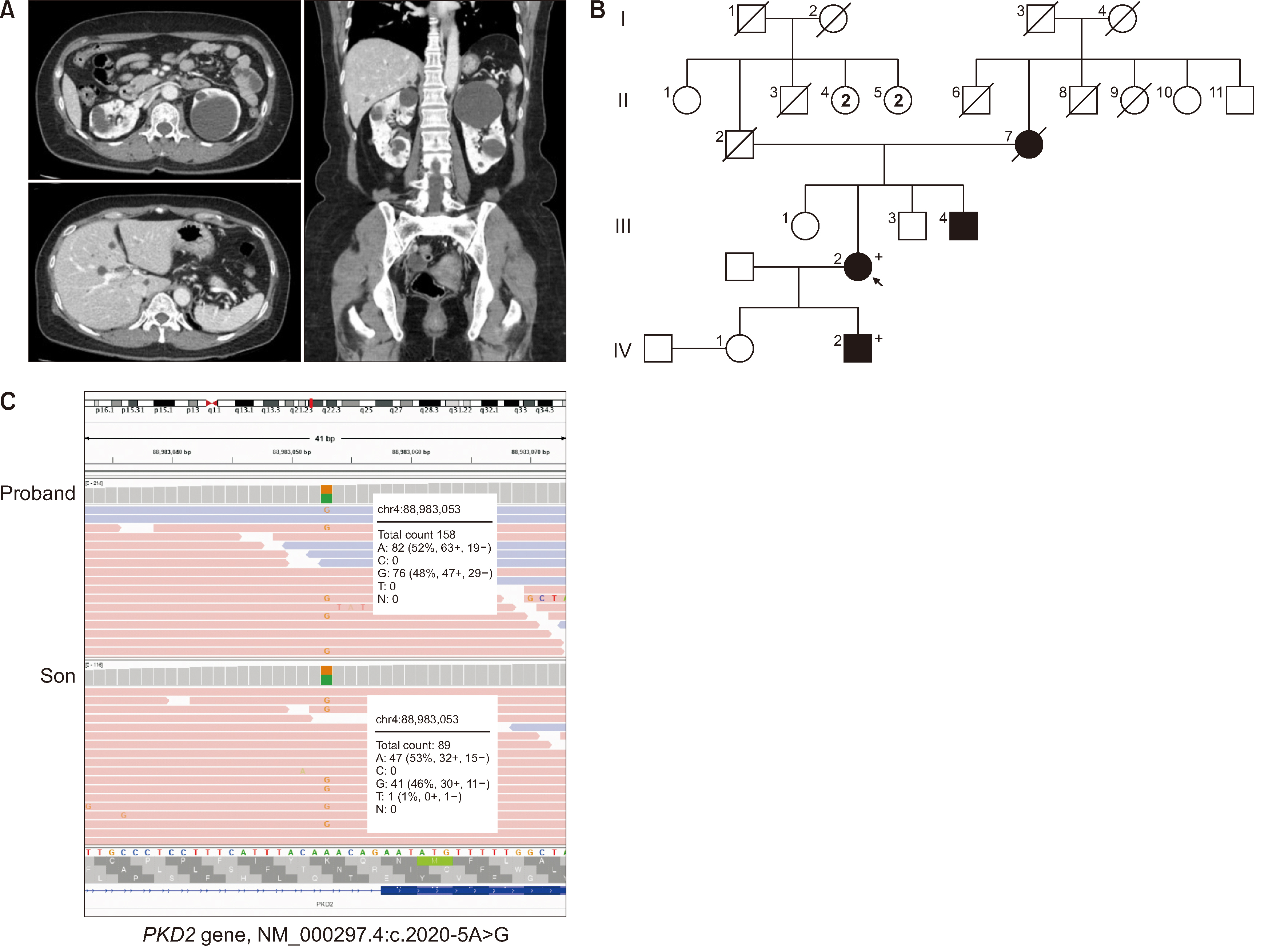
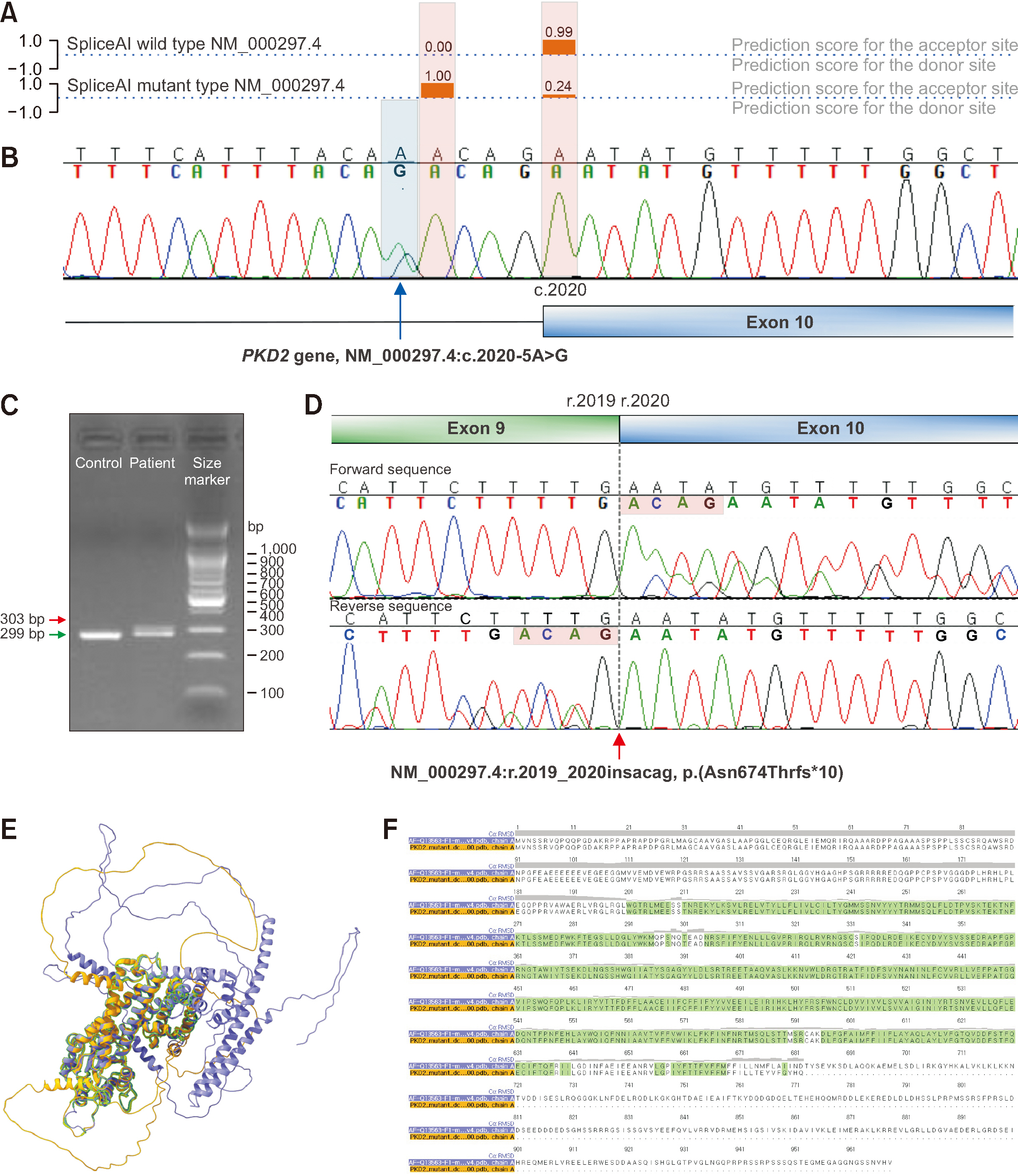
Aberrant Splicing in PKD2 in a Family of Korean Patients With Autosomal Dominant Polycystic Kidney Disease
- Affiliations
-
- 1Division of Nephrology, Department of Internal Medicine, Kyung Hee University College of Medicine, Kyung Hee University Medical Center, Seoul, Korea
- 2Department of Laboratory Medicine, Kyung Hee University College of Medicine, Kyung Hee University Medical Center, Seoul, Korea
- 3Rare Disease Center, Kyung Hee University College of Medicine, Kyung Hee University Medical Center, Seoul, Korea
- KMID: 2560816
- DOI: http://doi.org/10.3343/alm.2024.0221
Figure
Reference
-
References
1. Borrego Utiel FJ, Espinósa Hernandez M. 2023; How to estimate kidney growth in patients with autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 34:944–50. DOI: 10.1681/ASN.0000000000000130. PMID: 36995133. PMCID: PMC10278818.2. Lanktree MB, Haghighi A, di Bari I, Song X, Pei Y. 2021; Insights into autosomal dominant polycystic kidney disease from genetic studies. Clin J Am Soc Nephrol. 16:790–9. DOI: 10.2215/CJN.02320220. PMID: 32690722. PMCID: PMC8259493.3. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. 2019; Predicting splicing from primary sequence with deep learning. Cell. 176:535–48. e24. DOI: 10.1016/j.cell.2018.12.015. PMID: 30661751.4. Litchfield K, Reading JL, Lim EL, Xu H, Liu P, Al-Bakir M, et al. 2020; Escape from nonsense-mediated decay associates with anti-tumor immunogenicity. Nat Commun. 11:3800. DOI: 10.1038/s41467-020-17526-5. PMID: 32733040. PMCID: PMC7393139. PMID: 5361db97aec7487d9e4b4732b41ccecf.5. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. 2021; Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–9. DOI: 10.1038/s41586-021-03819-2. PMID: 34265844. PMCID: PMC8371605.6. Meng EC, Goddard TD, Pettersen EF, Couch GS, Pearson ZJ, Morris JH, et al. 2023; UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32:e4792. DOI: 10.1002/pro.4792. PMID: 37774136. PMCID: PMC10588335.7. Shen PS, Yang X, DeCaen PG, Liu X, Bulkley D, Clapham DE, et al. 2016; The structure of the polycystic kidney disease channel PKD2 in lipid nanodiscs. Cell. 167:763–73. e11. DOI: 10.1016/j.cell.2016.09.048. PMID: 27768895. PMCID: PMC6055481.8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.9. Walker LC, Hoya M, Wiggins GAR, Lindy A, Vincent LM, Parsons MT, et al. 2023; Using the ACMG/AMP framework to capture evidence related to predicted and observed impact on splicing: recommendations from the ClinGen SVI Splicing Subgroup. Am J Hum Genet. 110:1046–67. DOI: 10.1016/j.ajhg.2023.06.002. PMID: 37352859. PMCID: PMC10357475.10. Biesecker LG, Byrne AB, Harrison SM, Pesaran T, Schaffer AA, Shirts BH, et al. 2024; ClinGen guidance for use of the PP1/BS4 co-segregation and PP4 phenotype specificity criteria for sequence variant pathogenicity classification. Am J Hum Genet. 111:24–38. DOI: 10.1016/j.ajhg.2023.11.009. PMID: 38103548. PMCID: PMC10806742.11. Kim J, Yu J, Lee S, Kim S, Lee IS, Lee W, et al. 2024; Reverse transcription-PCR-based Sanger sequencing-confirmed exon-skipping effect of a novel GEN1 intronic variant (c.1408+4A>G). Ann Lab Med. 44:188–91. DOI: 10.3343/alm.2023.0163. PMID: 37840312. PMCID: PMC10628752.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Autosomal Dominant Polycystic Kidney Desease Coexisting with Renal Dysplasia. First Case Described and Followed Since Prenatal Period
- Clinical and genetic characteristics of Korean autosomal dominant polycystic kidney disease patients
- A Case of Renal Cell Carcinoma in Autosomal Dominant Polycystic Kidney Disease Hemodialyzed
- Differential Expression of PKD2-Associated Genes in Autosomal Dominant Polycystic Kidney Disease
- Autosomal Dominant Polycystic Kidney Desease Coexisting with Renal Dysplasia. First Case Described and Followed Since Prenatal Period