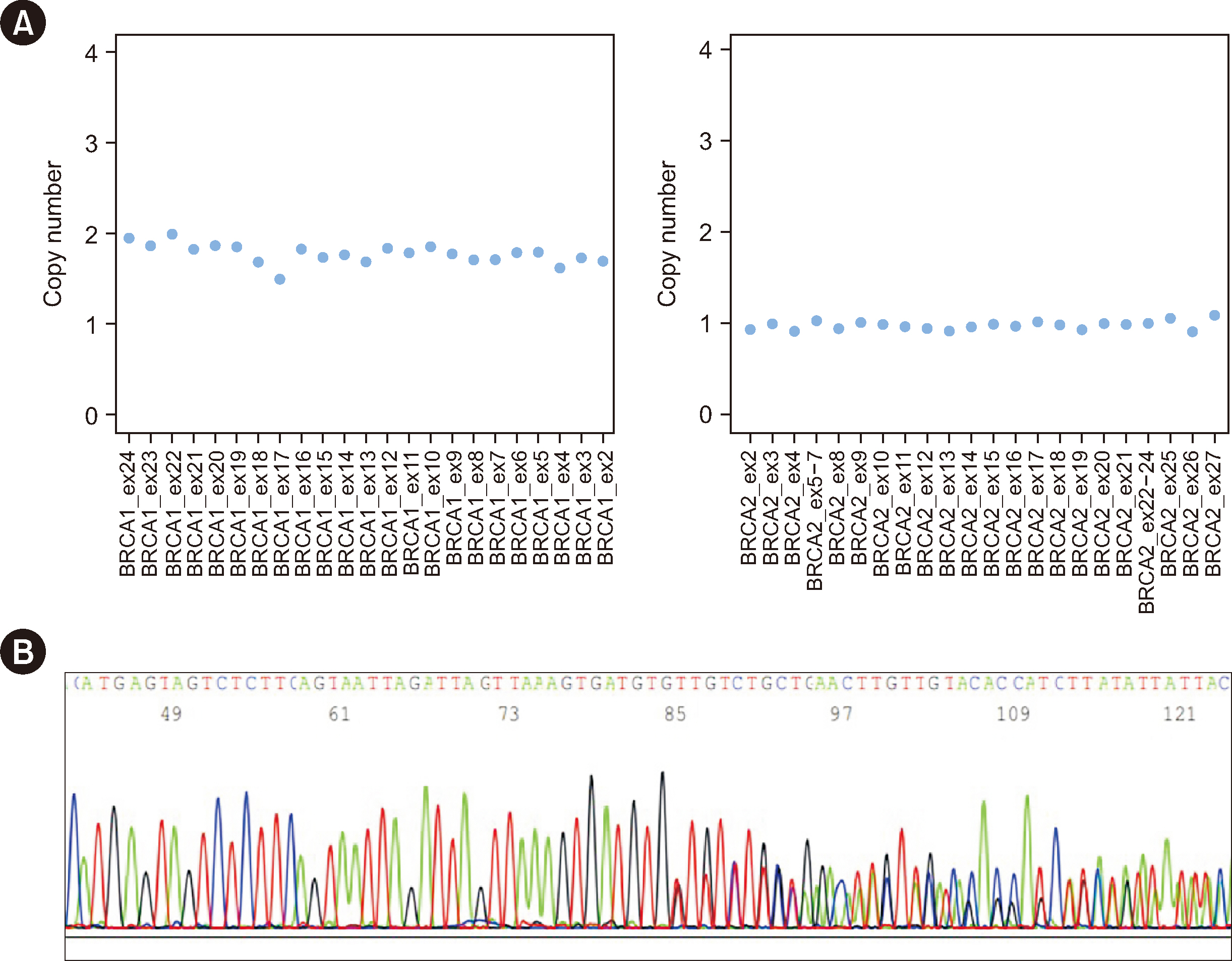
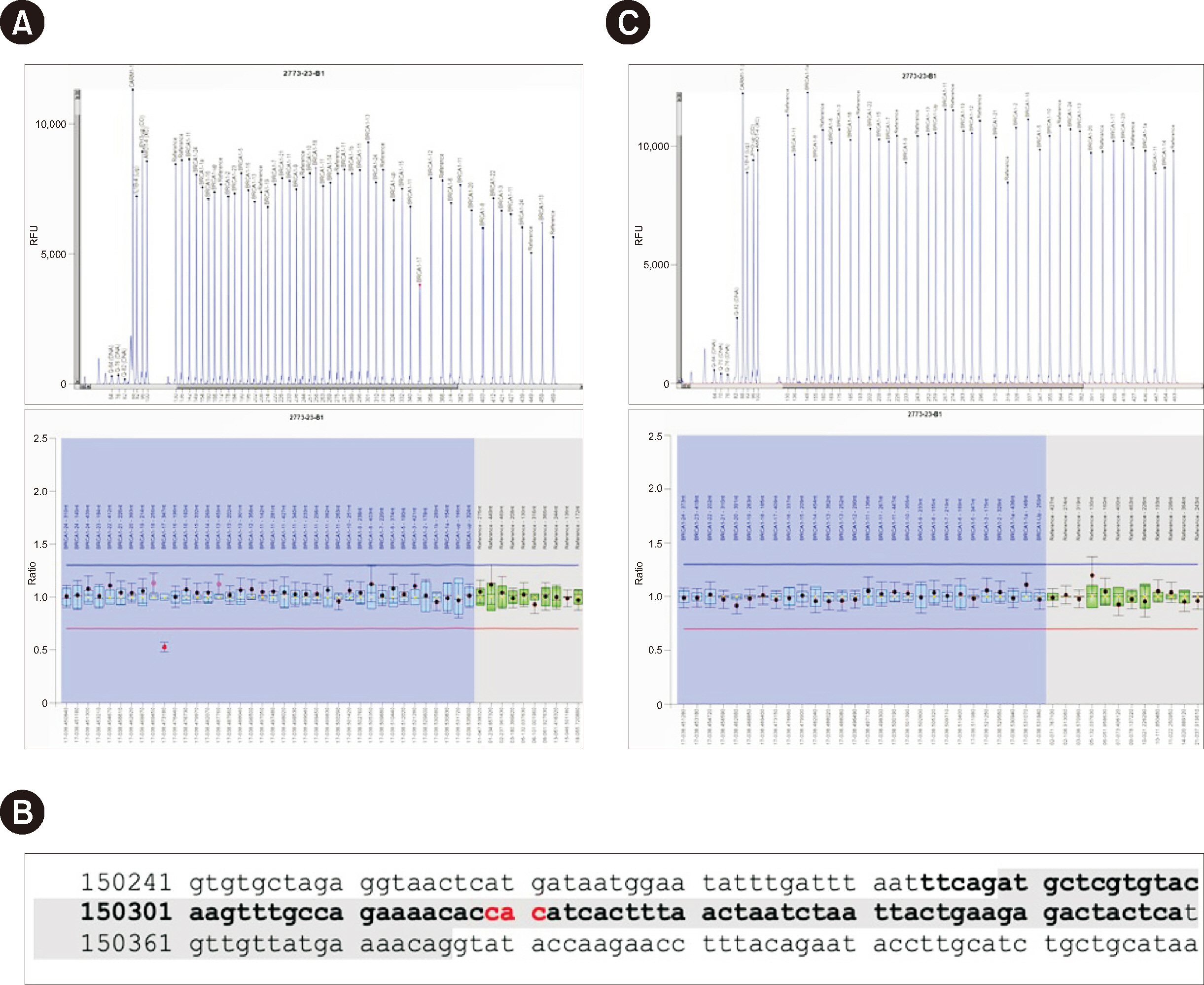
Ann Lab Med.
2024 Nov;44(6):617-620. 10.3343/alm.2024.0051.
Identification of a False-positive Multiplex Ligationdependent Probe Amplification Result in BRCA1 Using a Copy Number Variation Algorithm Under Development for a Commercial Next-Generation Sequencing-based Homologous Recombination Deficiency Assay
- Affiliations
-
- 1Departmental Unit of Molecular and Genomic Diagnostics, Policlinico Gemelli IRCCS Foundation, Rome, Italy
- 2Genomics Research Core Facility, Gemelli Science and Technology Park, Policlinico Gemelli IRCCS Foundation, Rome, Italy
- 3Genomics Research Core Facility, Gemelli Science and Technology Park, Policlinico Gemelli IRCCS Foundation, Rome, Italy
- 4Scientific Directorate, Policlinico Gemelli IRCCS Foundation, Rome, Italy
- 5Bioinformatics Research Core Facility, Gemelli Science and Technology Park, Policlinico Gemelli IRCCS Foundation, Rome, Italy
- 6Department of Clinical and Molecular Medicine, Faculty of Medicine and Psychology, Sapienza University of Rome, Rome, Italy
- 7S. Andrea University Hospital, Rome, Italy
- 8Institute of Obstetrics and Gynecology, Catholic University of the Sacred Heart, Rome, Italy
- KMID: 2560815
- DOI: http://doi.org/10.3343/alm.2024.0051
Figure
Reference
-
References
1. Kim KB, Park S, Ha JS, Ryoo N, Kim DH. 2022; Three cases of false-positive multiplex ligation-dependent probe amplification of BRCA1. Ann Lab Med. 42:497–9. DOI: 10.3343/alm.2022.42.4.497. PMID: 35177575. PMCID: PMC8859565.2. Cancer Genome Atlas Research Network. 2011; Integrated genomic analyses of ovarian carcinoma. Nature. 474:609–15. DOI: 10.1038/nature10166. PMID: 21720365. PMCID: PMC3163504.3. Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. 2015; Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 5:1137–54. DOI: 10.1158/2159-8290.CD-15-0714. PMID: 26463832. PMCID: PMC4631624.4. Giacò L, Palluzzi F, Guido D, Nero C, Giacomini F, Duranti S, et al. 2022; A computational framework for comprehensive genomic profiling in solid cancers: the analytical performance of a high-throughput assay for small and copy number variants. Cancers (Basel). 14:6152. DOI: 10.3390/cancers14246152. PMID: 36551638. PMCID: PMC9776229. PMID: 5fc382c2d0e5420191465cbf17292b1a.5. Minucci A, Scambia G, Santonocito C, Concolino P, Canu G, Mignone F, et al. 2015; Clinical impact on ovarian cancer patients of massive parallel sequencing for BRCA mutation detection: the experience at Gemelli hospital and a literature review. Expert Rev Mol Diagn. 15:1383–403. DOI: 10.1586/14737159.2015.1081059. PMID: 26306726.6. Concolino P, Mello E, Minucci A, Santonocito C, Scambia G, Giardina B, Capoluongo E. 2014; Advanced tools for BRCA1/2 mutational screening: comparison between two methods for large genomic rearrangements (LGRs) detection. Clin Chem Lab Med. 52:1119–27. DOI: 10.1515/cclm-2013-1114. PMID: 24670361.7. Minucci A, Scambia G, Santonocito C, Concolino P, Urbani A. 2020; BRCA testing in a genomic diagnostics referral center during the COVID-19 pandemic. Mol Biol Rep. 47:4857–60. DOI: 10.1007/s11033-020-05479-3. PMID: 32388698. PMCID: PMC7210797.8. Parsons MT, Tudini E, Li H, Hahnen E, Wappenschmidt B, Feliubadaló L, et al. 2019; Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum Mutat. 40:1557–78. DOI: 10.1002/humu.23818. PMID: 31131967. PMCID: PMC6772163.9. Zuntini R, Cortesi L, Calistri D, Pippucci T, Martelli PL, Casadio R, et al. 2014; BRCA1 p.His1673del is a pathogenic mutation associated with a predominant ovarian cancer phenotype. Oncotarget. 8:22640–8. DOI: 10.18632/oncotarget.15151. PMID: 28186987. PMCID: PMC5410251.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Determination of SMN1 and SMN2 Copy Numbers in a Korean Population using Multiplex Ligation-dependent Probe Amplification
- Performance Evaluation of BRCA1/2 Genetic Test Using Next-Generation Sequencing Based on Target Capture Method
- Diagnostic Effectiveness of Copy Number Variation Detection Using Multiplex Ligation-Dependent Probe Amplification in Patients with Lynch Syndrome-Related Cancer
- Utility of Next-Generation Sequencing Panel Including Hereditary Breast and Ovarian Cancer-Related Genes for Pathogenic Variant Detection
- Exonic copy number variations in rare genetic disorders