J Korean Med Sci.
2024 Oct;39(39):e271. 10.3346/jkms.2024.39.e271.
Identification of Preeclamptic Placenta in Whole Slide Images Using Artificial Intelligence Placenta Analysis
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Seoul National University College of Medicine, Seoul, Korea
- 2Department of Medical Device Development, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Transdisciplinary Medicine, Seoul National University Hospital, Seoul, Korea
- 4Interdisciplinary Program of Medical Informatics, Seoul National University College of Medicine, Seoul, Korea
- 5Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 6Department of Pathology, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul, Korea
- 7Department of Obstetrics and Gynecology, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul, Korea
- 8Department of Medicine, Seoul National University College of Medicine, Seoul, Korea
- 9Innovative Medical Technology Research Institute, Seoul National University Hospital, Seoul, Korea
- 10Department of Obstetrics and Gynecology, Seoul National University Hospital, Seoul, Korea
- 11Medical Big Data Research Center & Institute of Reproductive Medicine and Population, Medical Research Center, Seoul National University, Seoul, Korea
- KMID: 2560593
- DOI: http://doi.org/10.3346/jkms.2024.39.e271
Abstract
- Background
Preeclampsia (PE) is a hypertensive pregnancy disorder linked to placental dysfunction, often involving pathological lesions like acute atherosis, decidual vasculopathy, accelerated villous maturation, and fibrinoid deposition. However, there is no gold standard for the pathological diagnosis of PE and this limits the ability of clinicians to distinguish between PE and non-PE pregnancies. Recent advances in computational pathology have provided the opportunity to automate pathological analysis for diagnosis, classification, prediction, and prediction of disease progression. In this study, we assessed whether computational pathology could be used to identify PE placentas.
Methods
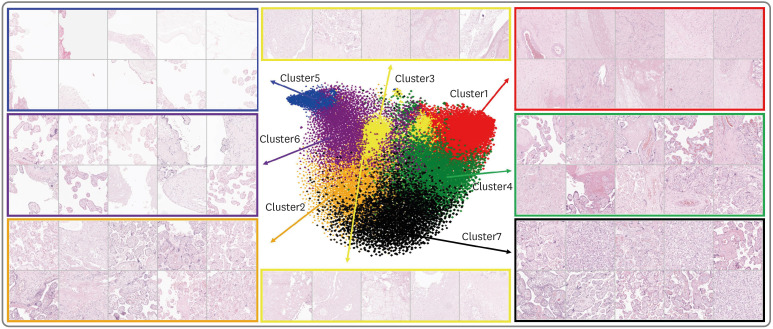
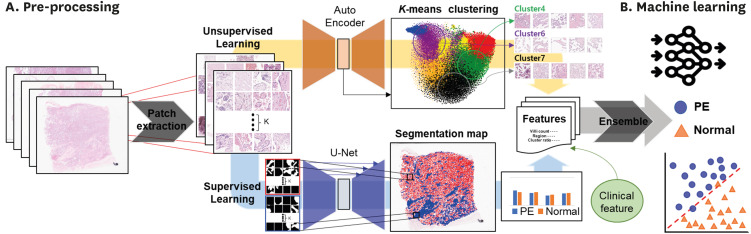
A total of 168 placental whole-slide images (WSIs) of patients from Seoul National University Hospital (comprising 84 PE cases and 84 normal controls) were used for model development and internal validation. For external validation of the model, 76 placental slides (including 38 PE cases and 38 normal controls) were obtained from the Boramae Medical Center (BMC). To establish standard criteria for diagnosing PE and distinguishing it from controls using placental WSIs, patch characteristics and quantification of terminal and intermediate villi were employed. In unsupervised learning, K-means clustering was conducted as a feature obtained through an Auto Encoder to extract the ratio of each cluster for each WSI. For supervised learning, quantitative assessments of the villi were obtained using a U-Net-based segmentation algorithm. The prediction model was developed using an ensemble method and was compared with a clinical feature model developed by using placental size features.
Results
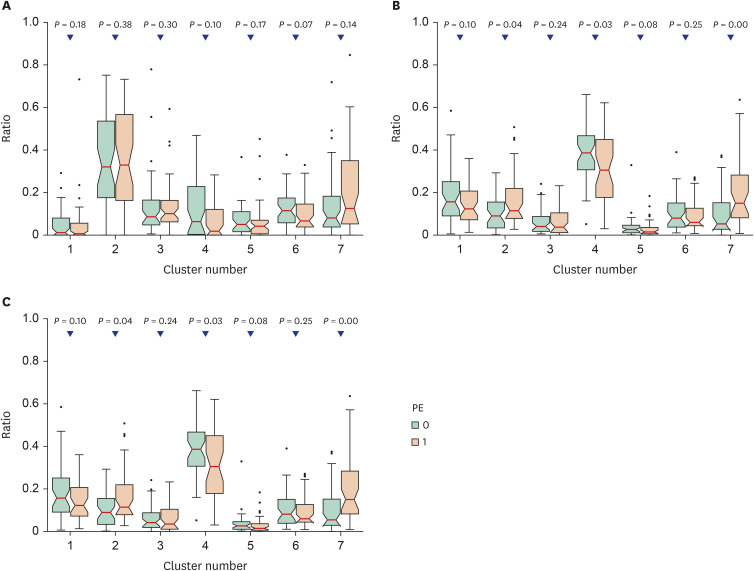
Using ensemble modeling, we developed a model to identify PE placentas. The model showed good performance (area under the precision-recall curve [AUPRC], 0.771; 95% confidence interval [CI], 0.752–0.790), with 77.3% of sensitivity and 71.1% of specificity, whereas the clinical feature model showed an AUPRC 0.713 (95% CI, 0.694–0.732) with 55.6% sensitivity and 86.8% specificity. External validation of the predictive model employing the BMC-derived set of placental slides also showed good discrimination (AUPRC, 0.725; 95% CI, 0.720–0.730).
Conclusion
The proposed computational pathology model demonstrated a strong ability to identify preeclamptic placentas. Computational pathology has the potential to improve the identification of PE placentas.
Figure
Reference
-
1. Abalos E, Cuesta C, Grosso AL, Chou D, Say L. Global and regional estimates of preeclampsia and eclampsia: a systematic review. Eur J Obstet Gynecol Reprod Biol. 2013; 170(1):1–7. PMID: 23746796.2. Say L, Chou D, Gemmill A, Tunçalp Ö, Moller AB, Daniels J, et al. Global causes of maternal death: a WHO systematic analysis. Lancet Glob Health. 2014; 2(6):e323–e333. PMID: 25103301.3. Ananth CV, Keyes KM, Wapner RJ. Pre-eclampsia rates in the United States, 1980-2010: age-period-cohort analysis. BMJ. 2013; 347:f6564. PMID: 24201165.4. Chappell LC, Cluver CA, Kingdom J, Tong S. Pre-eclampsia. Lancet. 2021; 398(10297):341–354. PMID: 34051884.5. Kulkarni VG, Sunilkumar KB, Nagaraj TS, Uddin Z, Ahmed I, Hwang K, et al. Maternal and fetal vascular lesions of malperfusion in the placentas associated with fetal and neonatal death: results of a prospective observational study. Am J Obstet Gynecol. 2021; 225(6):660.e1–660.e12.6. Roland CS, Hu J, Ren CE, Chen H, Li J, Varvoutis MS, et al. Morphological changes of placental syncytium and their implications for the pathogenesis of preeclampsia. Cell Mol Life Sci. 2016; 73(2):365–376. PMID: 26496726.7. Assibey-Mensah V, Parks WT, Gernand AD, Catov JM. Race and risk of maternal vascular malperfusion lesions in the placenta. Placenta. 2018; 69:102–108. PMID: 30213478.8. Li Z, Zhang J, Tan T, Teng X, Sun X, Zhao H, et al. Deep learning methods for lung cancer segmentation in whole-slide histopathology images-the ACDC@LungHP challenge 2019. IEEE J Biomed Health Inform. 2021; 25(2):429–440. PMID: 33216724.9. Khened M, Kori A, Rajkumar H, Krishnamurthi G, Srinivasan B. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci Rep. 2021; 11(1):11579. PMID: 34078928.10. Wang S, Yang DM, Rong R, Zhan X, Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol. 2019; 189(9):1686–1698. PMID: 31199919.11. Kanavati F, Toyokawa G, Momosaki S, Rambeau M, Kozuma Y, Shoji F, et al. Weakly-supervised learning for lung carcinoma classification using deep learning. Sci Rep. 2020; 10(1):9297. PMID: 32518413.12. Muhammad H, Sigel CS, Campanella G, Boerner T, Pak LM, Buttner S, et al. Unsupervised subtyping of cholangiocarcinoma using a deep clustering convolutional autoencoder. In : Proceedings of Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference; 2019 October 13–17; Shenzhen, China. Berlin, Germany: Springer International Publishing;2019.13. Bank D, Koenigstein N, Giryes R. Autoencoders. Rokach L, Maimon O, Shmueli E, editors. Machine Learning for Data Science Handbook: Data Mining and Knowledge Discovery Handbook. Berlin, Germany: Springer Nature;2023. p. 353–374.14. Kanungo T, Mount DM, Netanyahu NS, Piatko CD, Silverman R, Wu AY. An efficient k-means clustering algorithm: analysis and implementation. IEEE Trans Pattern Anal Mach Intell. 2002; 24(7):881–892.15. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In : Proceedings of Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference; 2015 October 5–9; Munich, Germany. Berlin, Germany: Springer;2015.16. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In : Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition; 2016 June 27–30; Las Vegas, NV, USA. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers (IEEE);2016.17. Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In : Proceedings of 2009 IEEE Computer Vision and Pattern Recognition; 2009 June 20–25; Miami, FL, USA. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers (IEEE);2009.18. Sarker IH. Machine learning: algorithms, real-world applications and research directions. SN Comput Sci. 2021; 2(3):160. PMID: 33778771.19. Prokhorenkova L, Gusev G, Vorobev A, Dorogush AV, Gulin A. CatBoost: unbiased boosting with categorical features. In : Proceedings of 32nd International Conference on Neural Information Processing Systems; 2018 December 3–8; Montreal, Canada. Red Hook, NY, USA: Curran Associates Inc.;2018.20. Boyd K, Eng KH, Page CD. Area under the precision-recall curve: point estimates and confidence intervals. In : Proceedings of Machine Learning and Knowledge Discovery in Databases: European Conference, ECML PKDD 2013; 2013 September 23–27; Prague, Czech Republic. Berlin, Germany: Springer;2013.21. Hossin M, Sulaiman MN. A review on evaluation metrics for data classification evaluations. Int J Data Min Knowl Manag Process. 2015; 5(2):1–11.22. Nelson DB, Ziadie MS, McIntire DD, Rogers BB, Leveno KJ. Placental pathology suggesting that preeclampsia is more than one disease. Am J Obstet Gynecol. 2014; 210(1):66.e1–66.e7.23. Hauspurg A, Redman EK, Assibey-Mensah V, Tony Parks W, Jeyabalan A, Roberts JM, et al. Placental findings in non-hypertensive term pregnancies and association with future adverse pregnancy outcomes: a cohort study. Placenta. 2018; 74:14–19. PMID: 30594310.24. Wang Q, Guo A. An efficient variance estimator of AUC and its applications to binary classification. Stat Med. 2020; 39(28):4281–4300. PMID: 32914457.25. Tharwat A.. Classification assessment methods. Appl Comput Inform. 2021; 17(1):168–192.26. Khong TY, Mooney EE, Ariel I, Balmus NC, Boyd TK, Brundler MA, et al. Sampling and definitions of placental lesions: Amsterdam Placental Workshop Group consensus statement. Arch Pathol Lab Med. 2016; 140(7):698–713. PMID: 27223167.27. Redline RW. Placental pathology: a systematic approach with clinical correlations. Placenta. 2008; 29(Suppl A):S86–S91. PMID: 17950457.28. Burton GJ, Woods AW, Jauniaux E, Kingdom JC. Rheological and physiological consequences of conversion of the maternal spiral arteries for uteroplacental blood flow during human pregnancy. Placenta. 2009; 30(6):473–482. PMID: 19375795.29. Pitz Jacobsen D, Fjeldstad HE, Johnsen GM, Fosheim IK, Moe K, Alnæs-Katjavivi P, et al. Acute atherosis lesions at the fetal-maternal border: current knowledge and implications for maternal cardiovascular health. Front Immunol. 2021; 12:791606. PMID: 34970270.30. Ashari IF, Dwi Nugroho E, Baraku R, Novri Yanda I, Liwardana R. Analysis of elbow, silhouette, Davies-Bouldin, Calinski-Harabasz, and rand-index evaluation on k-means algorithm for classifying flood-affected areas in Jakarta. J Appl Inform Comput. 2023; 7(1):95–103.31. Lee Y, Kim SY. Potential applications of ChatGPT in obstetrics and gynecology in Korea: a review article. Obstet Gynecol Sci. 2024; 67(2):153–159. PMID: 38247132.32. Ahn TG, Hwang JY. Preeclampsia and aspirin. Obstet Gynecol Sci. 2023; 66(3):120–132. PMID: 36924072.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Differential expression of Matrix Metalloproteinase (MMP)-2, -9 in normal and severe preeclamptic human placentas
- Expression of Placental Gelatinases in Normal and Preeclamptic Pregnancy
- Expression of 14-3-3 proteins and Bax in the placenta of preeclampsia
- Plasmin System in Placenta from Women with Normal and Preeclamptic Pregnancy
- A Case of Placenta Percreta Involving the Urinary Bladder