Cancer Res Treat.
2024 Oct;56(4):1027-1039. 10.4143/crt.2024.146.
Safe Utilization and Sharing of Genomic Data: Amendment to the Health and Medical Data Utilization Guidelines of South Korea
- Affiliations
-
- 1Department of Medical Sciences, Graduate School of The Catholic University of Korea, Seoul, Korea
- 2Department of Medical Informatics, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Department of Physiology, Ajou University School of Medicine, Suwon, Korea
- 4Center for Precision Medicine, Seoul National University Hospital, Seoul, Korea
- 5Department of Genomic Medicine, Seoul National University Hospital, Seoul, Korea
- 6Department of Life Science, Dongguk University, Seoul, Korea
- 7Department of Precision Medicine and Big Data, The Catholic University of Korea, Seoul, Korea
- 8Precision Medicine Research Center, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 9Cancer Evolution Research Center, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 10CMC Institute for Basic Medical Science, The Catholic Medical Center of The Catholic University of Korea, Seoul, Korea
- KMID: 2560237
- DOI: http://doi.org/10.4143/crt.2024.146
Abstract
- Purpose
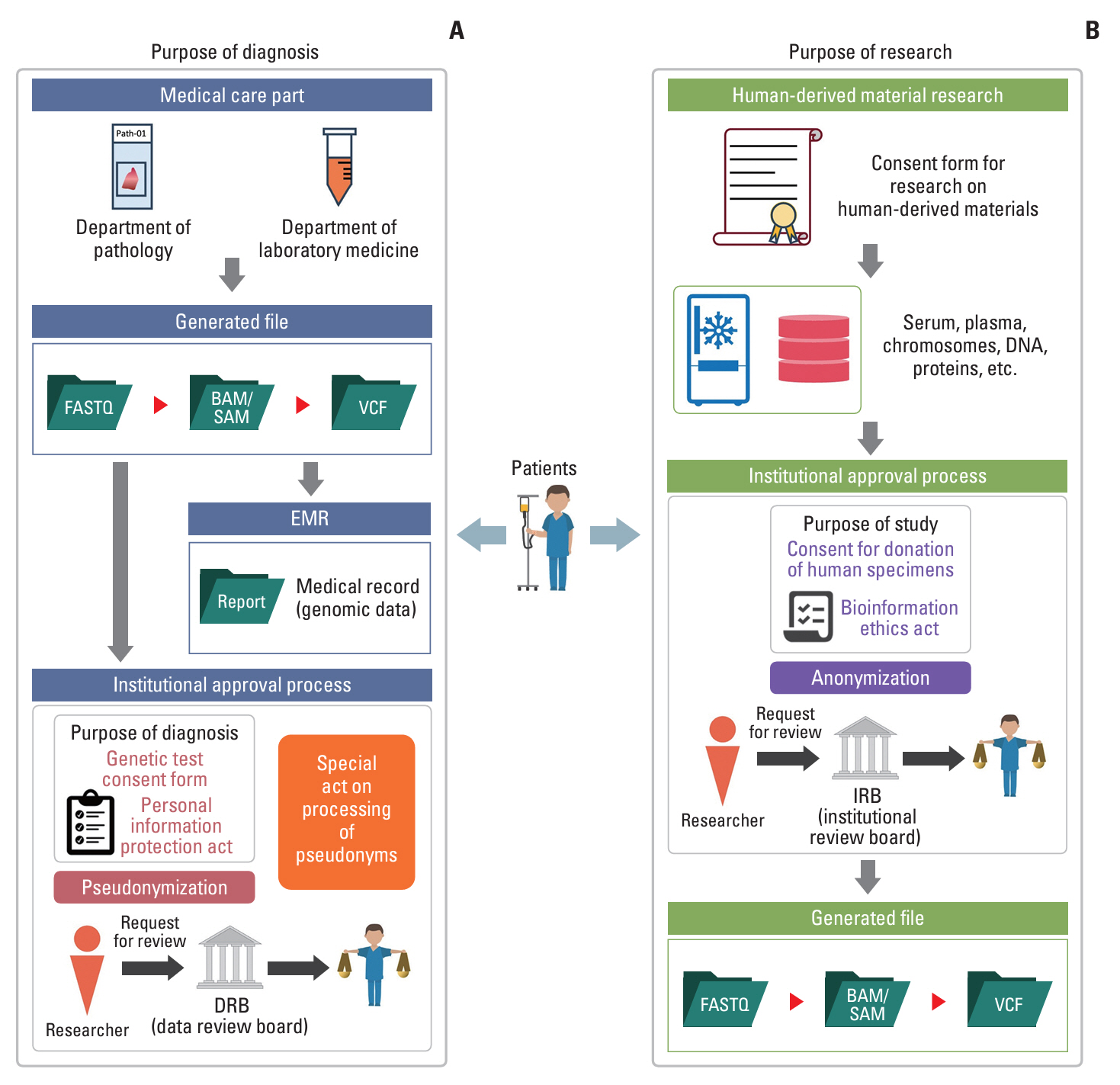
In 2024, medical researchers in the Republic of Korea were invited to amend the health and medical data utilization guidelines (Government Publications Registration Number: 11-1352000-0052828-14). This study aimed to show the overall impact of the guideline revision, with a focus on clinical genomic data.
Materials and Methods
This study amended the pseudonymization of genomic data defined in the previous version through a joint study led by the Ministry of Health and Welfare, the Korea Health Information Service, and the Korea Genome Organization. To develop the previous version, we held three conferences with four main medical research institutes and seven academic societies. We conducted two surveys targeting special genome experts in academia, industry, and institutes.
Results
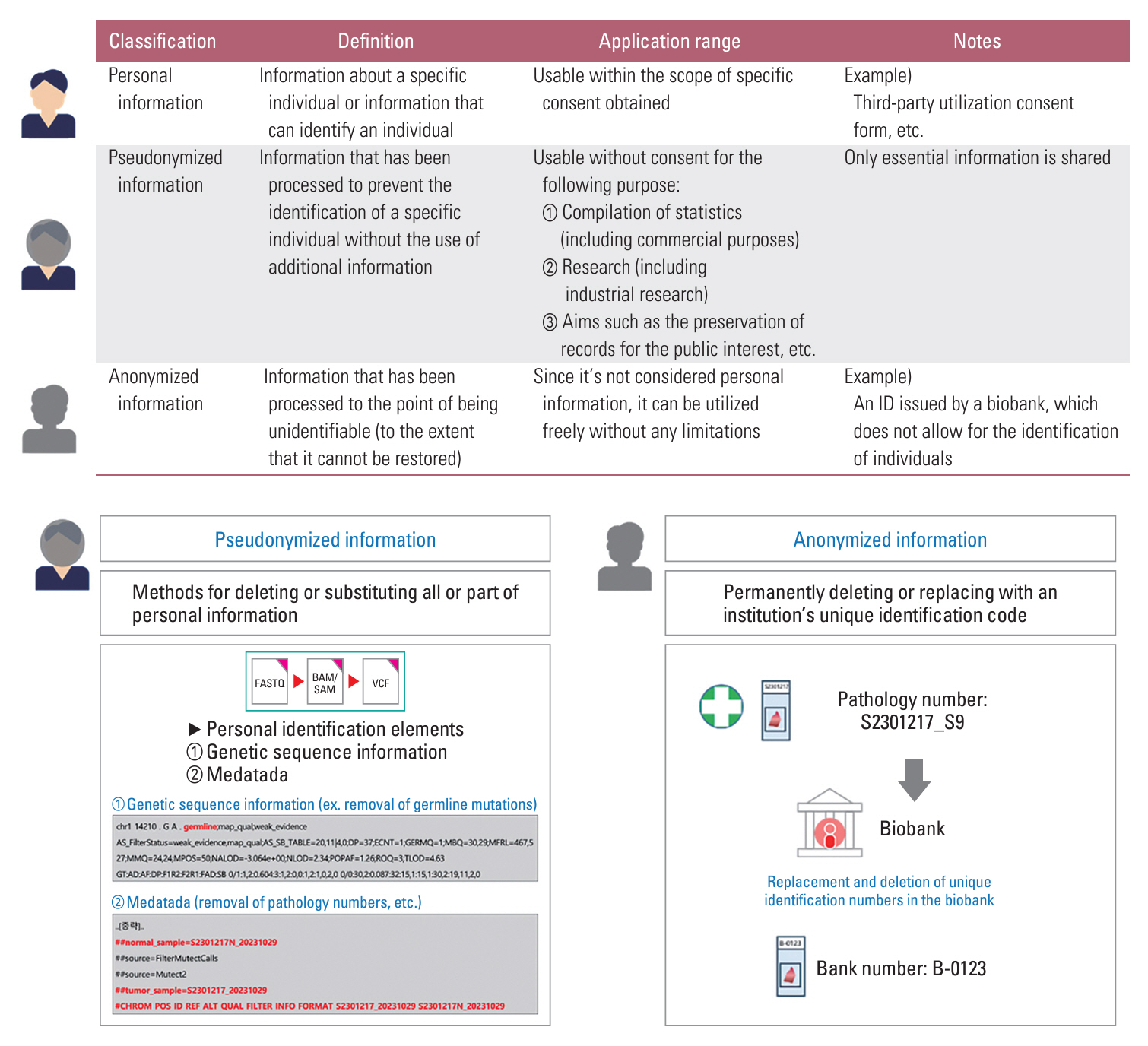
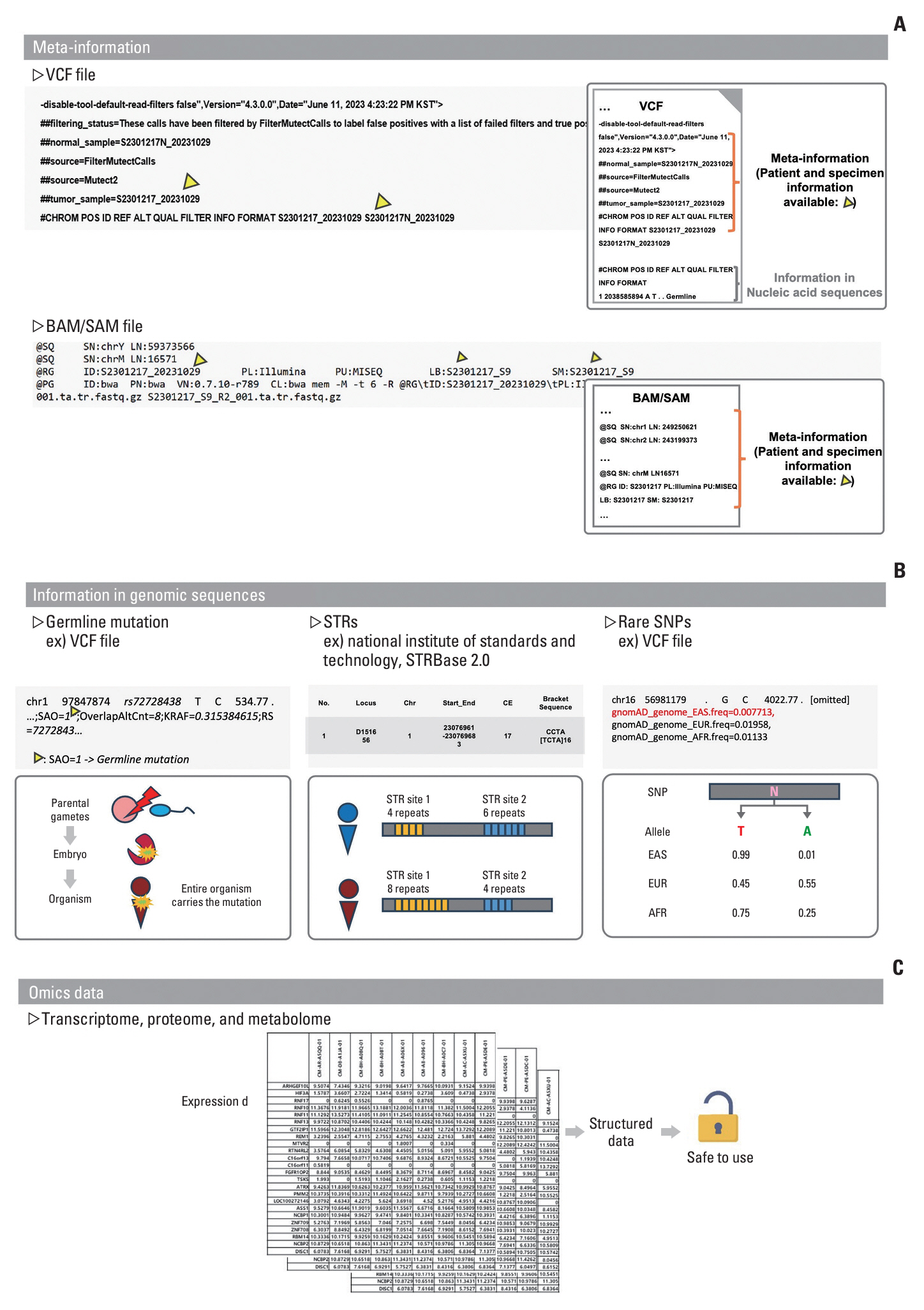
We found that cases of pseudonymization in the application of genome data were rare and that there was ambiguity in the terminology used in the previous version of the guidelines. Most experts (>~90%) agreed that the ‘reserved’ condition should be eliminated to make genomic data available after pseudonymization. In this study, the scope of genomic data was defined as clinical next-generation sequencing data, including FASTQ, BAM/SAM, VCF, and medical records. Pseudonymization targets genomic sequences and metadata, embedding specific elements, such as germline mutations, short tandem repeats, single-nucleotide polymorphisms, and identifiable data (for example, ID or environmental values). Expression data generated from multi-omics can be used without pseudonymization.
Conclusion
This amendment will not only enhance the safe use of healthcare data but also promote advancements in disease prevention, diagnosis, and treatment.
Keyword
Figure
Reference
-
References
1. Genomics market (by product and service: systems & software, consumables, services; by technology: sequencing, microarray, PCR, nucleic acid extraction and purification and others; by application: diagnostic application, drug discovery and development, agriculture and medical research and precision medicine and other; by end user: research institute, hospital and clinic, and others) - global industry analysis, size, share, growth, regional outlook and forecast 2023 to 2032 [Internet]. Ottawa: Precedence Research; c2023 [cited 2023 Feb 10]. Available from: https://www.precedenceresearch.com/genomics-market.2. Ramirez AH, Gebo KA, Harris PA. Progress with the All of Us Research Program: opening access for researchers. JAMA. 2021; 325:2441–2.3. All of Us Research Program Investigators, Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, et al. The “All of Us” Research Program. N Engl J Med. 2019; 381:668–76.
Article4. Ramirez AH, Sulieman L, Schlueter DJ, Halvorson A, Qian J, Ratsimbazafy F, et al. The All of Us Research Program: data quality, utility, and diversity. Patterns (N Y). 2022; 3:100570.5. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018; 562:203–9.
Article6. Collins R. What makes UK Biobank special? Lancet. 2012; 379:1173–4.
Article7. Littlejohns TJ, Holliday J, Gibson LM, Garratt S, Oesingmann N, Alfaro-Almagro F, et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat Commun. 2020; 11:2624.8. Lee B, Hwang S, Kim PG, Ko G, Jang K, Kim S, et al. Introduction of the Korea BioData Station (K-BDS) for sharing biological data. Genomics Inform. 2023; 21:e12.
Article9. Personal Information Protection Act [Internet]. Sejong: Korea Ministry of Government;2023. [cited 2023 Feb 10]. Available from: https://www.law.go.kr/LSW//lsInfoP.do?lsiSeq=213857&chrClsCd=010203&urlMode=engLsInfoR&viewCls=engLsInfoR#0000.10. Kim YJ, Moon S, Hwang MY, Han S, Jang HM, Kong J, et al. The contribution of common and rare genetic variants to variation in metabolic traits in 288,137 East Asians. Nat Commun. 2022; 13:6642.
Article11. Kim Y, Han BG; KoGES Group. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) Consortium. Int J Epidemiol. 2017; 46:e20.
Article12. Jeon Y, Jeon S, Blazyte A, Kim YJ, Lee JJ, Bhak Y, et al. Welfare genome project: a participatory Korean Personal Genome Project with free health check-up and genetic report followed by counseling. Front Genet. 2021; 12:633731.
Article13. Enforcement Rule of Bioethics and Safety Act [Internet]. Sejong: Korea Ministry of Government;2023. [cited 2023 Feb 10]. Available from: https://www.law.go.kr/LSW/lsInfoP.do?lsiSeq=98198&urlMode=engLsInfoR&viewCls=engLsInfoR#0000.14. Edge MD, Algee-Hewitt BF, Pemberton TJ, Li JZ, Rosenberg NA. Linkage disequilibrium matches forensic genetic records to disjoint genomic marker sets. Proc Natl Acad Sci U S A. 2017; 114:5671–6.
Article15. Carter AB. Considerations for genomic data privacy and security when working in the cloud. J Mol Diagn. 2019; 21:542–52.
Article16. Phillips C, Prieto L, Fondevila M, Salas A, Gomez-Tato A, Alvarez-Dios J, et al. Ancestry analysis in the 11-M Madrid bomb attack investigation. PLoS One. 2009; 4:e6583.
Article17. Sanchez JJ, Phillips C, Borsting C, Balogh K, Bogus M, Fondevila M, et al. A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis. 2006; 27:1713–24.
Article18. Pakstis AJ, Speed WC, Fang R, Hyland FC, Furtado MR, Kidd JR, et al. SNPs for a universal individual identification panel. Hum Genet. 2010; 127:315–24.
Article19. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–60.
Article20. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–303.
Article21. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–9.
Article22. Jeon S, Choi H, Jeon Y, Choi WH, Choi H, An K, et al. Korea4K: whole genome sequences of 4,157 Koreans with 107 phenotypes derived from extensive health check-ups. Gigascience. 2024; 13:giae014.
Article23. Kuo TT, Jiang X, Tang H, Wang X, Bath T, Bu D, et al. iDASH secure genome analysis competition 2018: blockchain genomic data access logging, homomorphic encryption on GWAS, and DNA segment searching. BMC Med Genomics. 2020; 13(Suppl 7):98.
Article24. Raisaro JL, Gwangbae C, Pradervand S, Colsenet R, Jacquemont N, Rosat N, et al. Protecting privacy and security of genomic data in i2b2 with homomorphic encryption and differential privacy. IEEE/ACM Trans Comput Biol Bioinform. 2018; 15:1413–26.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Effects of Case Management for Medicaid on Healthcare Utilization by the Medicaid System
- The Effect of Outpatient Cost Sharing on Health Care Utilization of the Elderly
- Determinants Influencing the Utilization of the Rural Health Sub-centers
- Factors associated with health services utilization between the years 2010 and 2012 in Korea: using Andersen's Behavioral model
- Changes in Health Care Utilization during the COVID-19 Pandemic