Ann Lab Med.
2024 Jul;44(4):324-334. 10.3343/alm.2023.0339.
Comparison of Optical Genome Mapping With Conventional Diagnostic Methods for Structural Variant Detection in Hematologic Malignancies
- Affiliations
-
- 1Brain Korea 21 PLUS Project for Medical Science, Yonsei University, Seoul, Korea
- 2MDxK (Molecular Diagnostics Korea), Inc., Gwacheon, Korea
- 3Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea; 4 Dxome Co., Ltd., Seongnam, Korea
- KMID: 2557931
- DOI: http://doi.org/10.3343/alm.2023.0339
Abstract
- Background
Structural variants (SVs) are currently analyzed using a combination of conventional methods; however, this approach has limitations. Optical genome mapping (OGM), an emerging technology for detecting SVs using a single-molecule strategy, has the potential to replace conventional methods. We compared OGM with conventional diagnostic methods for detecting SVs in various hematologic malignancies.
Methods
Residual bone marrow aspirates from 27 patients with hematologic malignancies in whom SVs were observed using conventional methods (chromosomal banding analysis, FISH, an RNA fusion panel, and reverse transcription PCR) were analyzed using OGM. The concordance between the OGM and conventional method results was evaluated.
Results
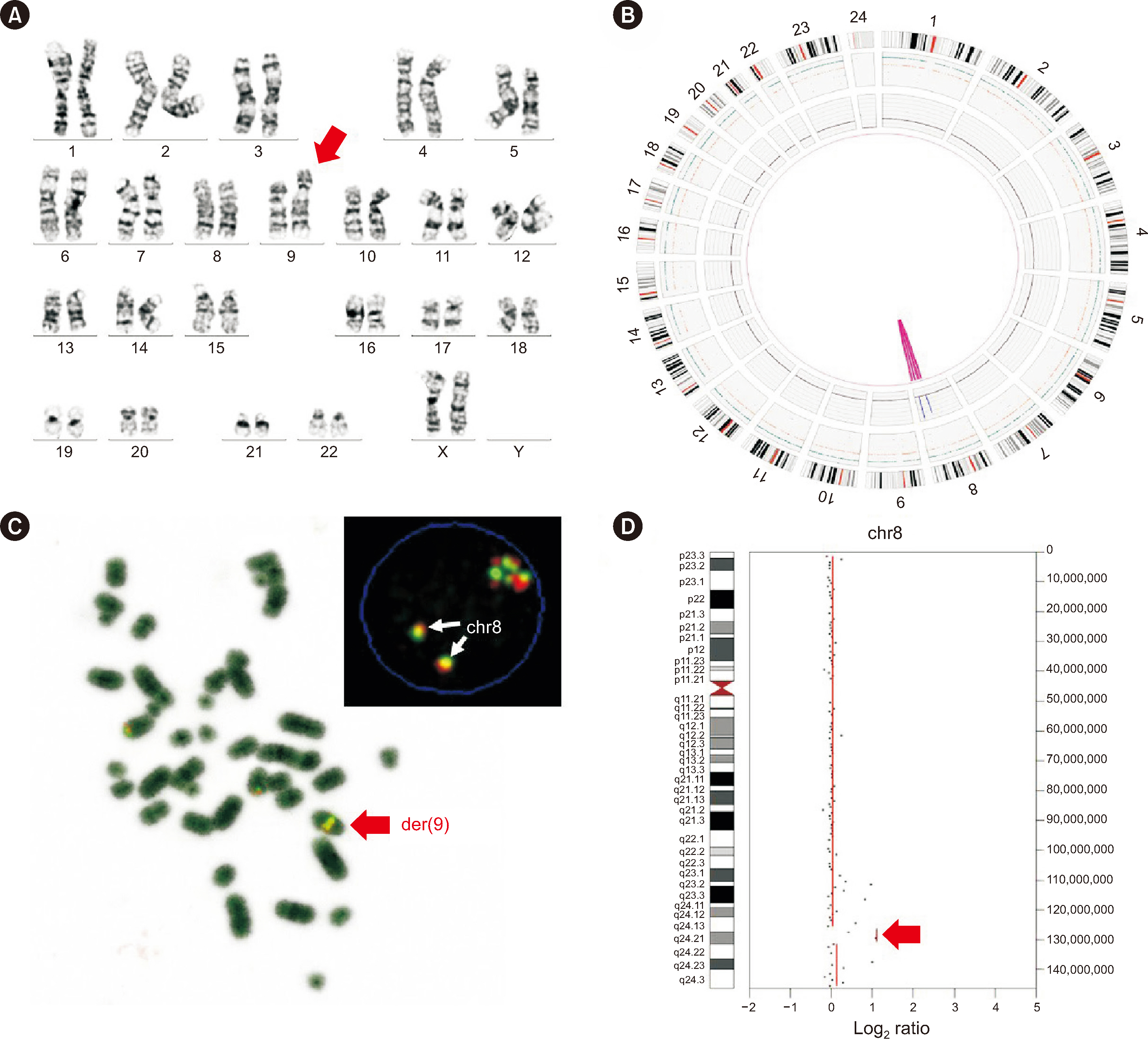
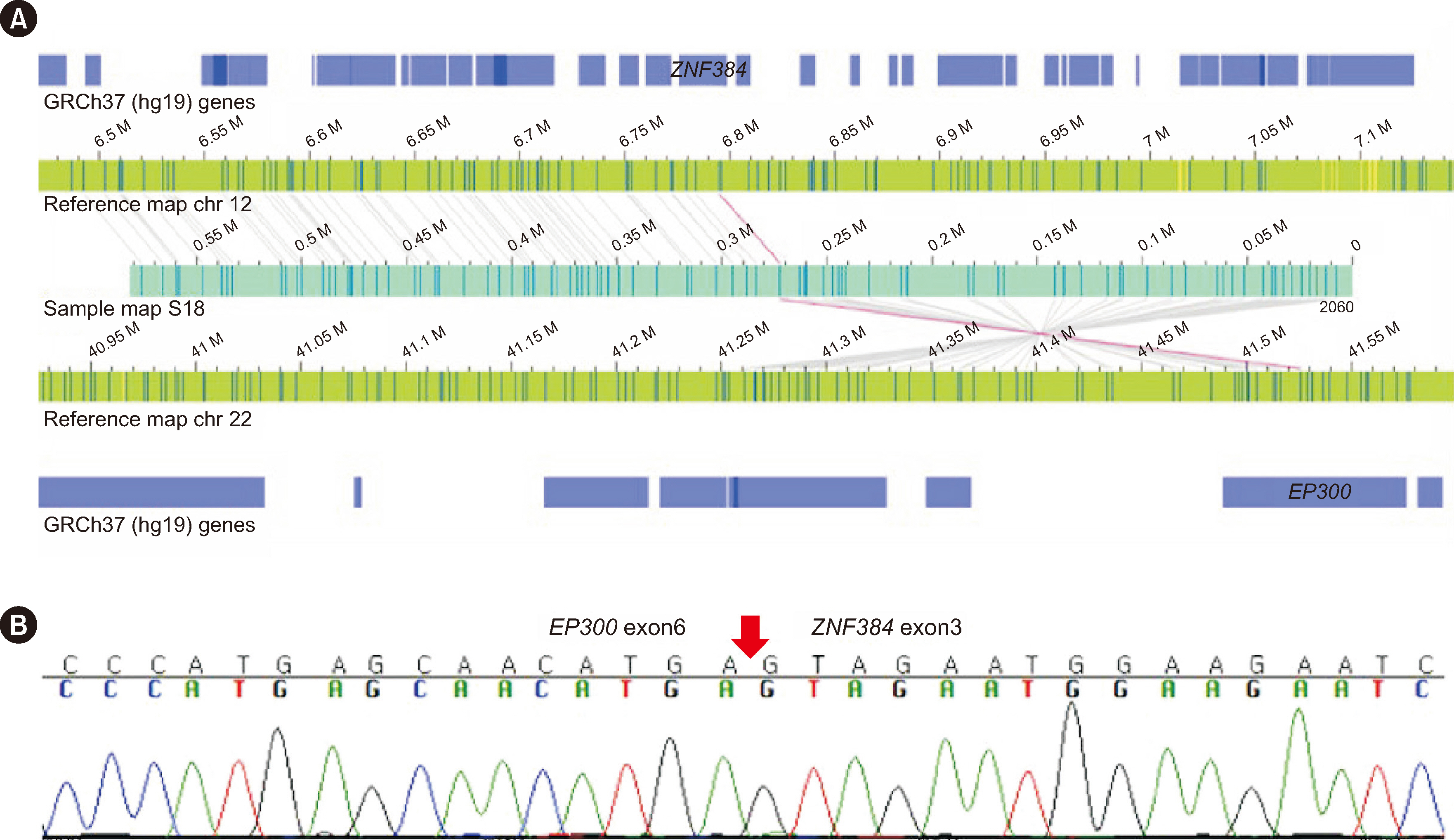
OGM showed concordance in 63% (17/27) and partial concordance in 37% (10/27) of samples. OGM detected 76% (52/68) of the total SVs correctly (concordance rate for each type of SVs: aneuploidies, 83% [15/18]; balanced translocation, 80% [12/15] unbalanced translocation, 54% [7/13] deletions, 81% [13/16]; duplications, 100% [2/2] inversion 100% [1/1]; insertion, 100% [1/1]; marker chromosome, 0% [0/1]; isochromosome, 100% [1/1]). Sixteen discordant results were attributed to the involvement of centromeric/telomeric regions, detection sensitivity, and a low mapping rate and coverage. OGM identified additional SVs, including submicroscopic SVs and novel fusions, in five cases.
Conclusions
OGM shows a high level of concordance with conventional diagnostic methods for the detection of SVs and can identify novel variants, suggesting its potential utility in enabling more comprehensive SV analysis in routine diagnostics of hematologic malignancies, although further studies and improvements are required.
Keyword
Figure
Cited by 1 articles
-
Application of Optical Genome Mapping to the Genetic Diagnosis of Facioscapulohumeral Muscular Dystrophy 1
Seung-Tae Lee
Ann Lab Med. 2024;44(5):383-384. doi: 10.3343/alm.2024.0197.
Reference
-
References
1. Schütte J, Reusch J, Khandanpour C, Eisfeld C. 2019; Structural variants as a basis for targeted therapies in hematological malignancies. Front Oncol. 9:839. DOI: 10.3389/fonc.2019.00839. PMID: 31555592. PMCID: PMC6722867. PMID: 2316a34d446d40e1b692af6b6e461ccd.
Article2. Park MS, Kim HY, Lee JJ, Cho D, Jung CW, Kim HJ, et al. 2023; The first case of acute myeloid leukemia with t(10;11)(p13;q21);PICALM-MLLT10 rearrangement presenting with extensive skin involvement. Ann Lab Med. 43:310–4. DOI: 10.3343/alm.2023.43.3.310. PMID: 36544346. PMCID: PMC9791006.
Article3. Ho SS, Urban AE, Mills RE. 2020; Structural variation in the sequencing era. Nat Rev Genet. 21:171–89. DOI: 10.1038/s41576-019-0180-9. PMID: 31729472. PMCID: PMC7402362.
Article4. Braggio E, Egan JB, Fonseca R, Stewart AK. 2013; Lessons from next-generation sequencing analysis in hematological malignancies. Blood Cancer J. 3:e127. DOI: 10.1038/bcj.2013.26. PMID: 23872706. PMCID: PMC3730204.
Article5. Merker JD, Valouev A, Gotlib J. 2012; Next-generation sequencing in hematologic malignancies: what will be the dividends? Ther Adv Hematol. 3:333–9. DOI: 10.1177/2040620712458948. PMID: 23606936. PMCID: PMC3627325.
Article6. He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, et al. 2016; Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 127:3004–14. DOI: 10.1182/blood-2015-08-664649. PMID: 26966091. PMCID: PMC4968346.
Article7. Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, et al. 2018; Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 36:338–45. DOI: 10.1038/nbt.4060. PMID: 29431738. PMCID: PMC5889714.
Article8. Feuk L, Carson AR, Scherer SW. 2006; Structural variation in the human genome. Nat Rev Genet. 7:85–97. DOI: 10.1038/nrg1767. PMID: 16418744.
Article9. Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, et al. 2012; Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 30:771–6. DOI: 10.1038/nbt.2303. PMID: 22797562. PMCID: PMC3817024.
Article10. Hastie AR, Dong L, Smith A, Finklestein J, Lam ET, Huo N, et al. 2013; Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLoS One. 8:e55864. DOI: 10.1371/journal.pone.0055864. PMID: 23405223. PMCID: PMC3566107. PMID: c7b413d434f142c2b1a2121f0325848e.
Article11. Cao H, Hastie AR, Cao D, Lam ET, Sun Y, Huang H, et al. 2014; Rapid detection of structural variation in a human genome using nanochannel-based genome mapping technology. Gigascience. 3:34. DOI: 10.1186/2047-217X-3-34. PMID: 25671094. PMCID: PMC4322599.
Article12. Chan S, Lam E, Saghbini M, Bocklandt S, Hastie A, Cao H, et al. 2018; Structural variation detection and analysis using Bionano optical mapping. Methods Mol Biol. 1833:193–203. DOI: 10.1007/978-1-4939-8666-8_16. PMID: 30039375.
Article13. Neveling K, Mantere T, Vermeulen S, Oorsprong M, van Beek R, Kater-Baats E, et al. 2021; Next-generation cytogenetics: comprehensive assessment of 52 hematological malignancy genomes by optical genome mapping. Am J Hum Genet. 108:1423–35. DOI: 10.1016/j.ajhg.2021.06.001. PMID: 34237281. PMCID: PMC8387283.
Article14. Rack K, De Bie J, Ameye G, Gielen O, Demeyer S, Cools J, et al. 2022; Optimizing the diagnostic workflow for acute lymphoblastic leukemia by optical genome mapping. Am J Hematol. 97:548–61. DOI: 10.1002/ajh.26487. PMID: 35119131. PMCID: PMC9314940.
Article15. Gerding WM, Tembrink M, Nilius-Eliliwi V, Mika T, Dimopoulos F, Ladigan-Badura S, et al. 2022; Optical genome mapping reveals additional prognostic information compared to conventional cytogenetics in AML/MDS patients. Int J Cancer. 150:1998–2011. DOI: 10.1002/ijc.33942. PMID: 35064925.
Article16. Puiggros A, Ramos-Campoy S, Kamaso J, de la Rosa M, Salido M, Melero C, et al. 2022; Optical genome mapping: a promising new tool to assess genomic complexity in chronic lymphocytic leukemia (CLL). Cancers (Basel). 14:DOI: 10.3390/cancers14143376. PMID: 35884436. PMCID: PMC9317182. PMID: 832f49ffea7a48748c6993b7381de703.
Article17. Soler G, Ouedraogo ZG, Goumy C, Lebecque B, Aspas Requena G, Ravinet A, et al. 2023; Optical genome mapping in routine cytogenetic diagnosis of acute leukemia. Cancers (Basel). 15:DOI: 10.3390/cancers15072131. PMID: 37046792. PMCID: PMC10093111. PMID: a6efa1b02a1e4a7785e327076b3f30da.
Article18. Lestringant V, Duployez N, Penther D, Luquet I, Derrieux C, Lutun A, et al. 2021; Optical genome mapping, a promising alternative to gold standard cytogenetic approaches in a series of acute lymphoblastic leukemias. Genes Chromosomes Cancer. 60:657–67. DOI: 10.1002/gcc.22971. PMID: 33982372.
Article19. Sahajpal NS, Mondal AK, Tvrdik T, Hauenstein J, Shi H, Deeb KK, et al. 2022; Clinical validation and diagnostic utility of optical genome mapping for enhanced cytogenomic analysis of hematological neoplasms. J Mol Diagn. 24:1279–91. DOI: 10.1016/j.jmoldx.2022.09.009. PMID: 36265723.20. Brandes D, Yasin L, Nebral K, Ebler J, Schinnerl D, Picard D, et al. 2023; Optical genome mapping identifies novel recurrent structural alterations in childhood ETV6::RUNX1+ and high hyperdiploid acute lymphoblastic leukemia. Hemasphere. 7:e925. DOI: 10.1097/HS9.0000000000000925. PMID: 37469802. PMCID: PMC10353714. PMID: 2b7eb25f131c4839b43dc5acad546223.21. Yang H, Garcia-Manero G, Sasaki K, Montalban-Bravo G, Tang Z, Wei Y, et al. 2022; High-resolution structural variant profiling of myelodysplastic syndromes by optical genome mapping uncovers cryptic aberrations of prognostic and therapeutic significance. Leukemia. 36:2306–16. DOI: 10.1038/s41375-022-01652-8. PMID: 35915143. PMCID: PMC9417987.
Article22. Shaffer LG, McGowan-Jordan J, Schmid M, editors. 2013. ISCN 2013: an international system for human cytogenetic nomenclature (2013). Karger Medical and Scientific Publishers;Basel: DOI: 10.1002/ajmg.a.35995.23. Kim B, Kim E, Lee ST, Cheong JW, Lyu CJ, Min YH, et al. 2020; Detection of recurrent, rare, and novel gene fusions in patients with acute leukemia using next-generation sequencing approaches. Hematol Oncol. 38:82–8. DOI: 10.1002/hon.2709. PMID: 31875988.
Article24. Kim B, Lee H, Shin S, Lee ST, Choi JR. 2019; Clinical evaluation of massively parallel RNA sequencing for detecting recurrent gene fusions in hematologic malignancies. J Mol Diagn. 21:163–70. DOI: 10.1016/j.jmoldx.2018.09.002. PMID: 30347268.
Article25. Kim B, Lee H, Kim E, Shin S, Lee ST, Choi JR. 2019; Clinical utility of targeted NGS panel with comprehensive bioinformatics analysis for patients with acute lymphoblastic leukemia. Leuk Lymphoma. 60:3138–45. DOI: 10.1080/10428194.2019.1627538. PMID: 31203682.
Article26. Kim B, Lee H, Jang J, Kim SJ, Lee ST, Cheong JW, et al. 2019; Targeted next generation sequencing can serve as an alternative to conventional tests in myeloid neoplasms. PLoS One. 14:e0212228. DOI: 10.1371/journal.pone.0212228. PMID: 30840646. PMCID: PMC6402635. PMID: ebbfbd014c8945768ae040b5eb346d60.
Article27. Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, et al. 2012; A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 28:2747–54. DOI: 10.1093/bioinformatics/bts526. PMID: 22942019. PMCID: PMC3476336.
Article28. Kim H, Shim Y, Lee TG, Won D, Choi JR, Shin S, et al. 2023; Copy-number analysis by base-level normalization: An intuitive visualization tool for evaluating copy number variations. Clin Genet. 103:35–44. DOI: 10.1111/cge.14236. PMID: 36152294.
Article29. Kuilman T, Velds A, Kemper K, Ranzani M, Bombardelli L, Hoogstraat M, et al. 2015; CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 16:49. DOI: 10.1186/s13059-015-0617-1. PMID: 25887352. PMCID: PMC4396974.
Article30. Nilius-Eliliwi V, Gerding WM, Schroers R, Nguyen HP, Vangala DB. 2023; Optical genome mapping for cytogenetic diagnostics in AML. Cancers (Basel). 15:DOI: 10.3390/cancers15061684. PMID: 36980569. PMCID: PMC10046241. PMID: 8044125cd05145fa8bb2bea74e9670dd.
Article31. Wang Y, Li J, Xue TL, Tian S, Yue ZX, Liu SG, et al. 2023; Clinical, biological, and outcome features of P2RY8-CRLF2 and CRLF2 over-expression in pediatric B-cell precursor acute lymphoblastic leukemia according to the CCLG-ALL 2008 and 2018 protocol. Eur J Haematol. 110:669–79. DOI: 10.1111/ejh.13948. PMID: 36814093.32. Palmi C, Vendramini E, Silvestri D, Longinotti G, Frison D, Cario G, et al. 2012; Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia. 26:2245–53. DOI: 10.1038/leu.2012.101. PMID: 22484421.
Article33. Cario G, Zimmermann M, Romey R, Gesk S, Vater I, Harbott J, et al. 2010; Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 115:5393–7. DOI: 10.1182/blood-2009-11-256131. PMID: 20378752.
Article34. Ahmadi SE, Rahimi S, Zarandi B, Chegeni R, Safa M. 2021; MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J Hematol Oncol. 14:121. DOI: 10.1186/s13045-021-01111-4. PMID: 34372899. PMCID: PMC8351444. PMID: 9225a1d6c9c74a6797548088c0548e98.
Article35. Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. 2017; ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 102:118–29. DOI: 10.3324/haematol.2016.151035. PMID: 27634205. PMCID: PMC5210242.36. Skucha A, Ebner J, Grebien F. 2019; Roles of SETD2 in leukemia-transcription, DNA-damage, and beyond. Int J Mol Sci. 20:DOI: 10.3390/ijms20051029. PMID: 30818762. PMCID: PMC6429614. PMID: 38ce543530fe49fca74514bffcd2f4d4.
Article37. Vairy S, Tran TH. 2020; IKZF1 alterations in acute lymphoblastic leukemia: the good, the bad and the ugly. Blood Rev. 44:100677. DOI: 10.1016/j.blre.2020.100677. PMID: 32245541.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Application of Optical Genome Mapping to the Genetic Diagnosis of Facioscapulohumeral Muscular Dystrophy 1
- Genomic technologies for detecting structural variations in hematologic malignancies
- Korean Society for Genetic Diagnostics Guidelines for Validation of Next-Generation Sequencing-Based Somatic Variant Detection in Hematologic Malignancies
- Automatic detection of tooth cracks in optical coherence tomography images
- Clinically relevant core genes for hematologic malignancies in clinical NGS panel testing