J Korean Med Sci.
2024 Jul;39(27):e198. 10.3346/jkms.2024.39.e198.
Validation of the Utility of the Genetically Shared Regions of Chromosomes (GD-ICS) Measuring Method in Identifying Complicated Genetic Relatedness
- Affiliations
-
- 1Institute of Forensic and Anthropological Science, Seoul National University Medical Research Center, Seoul, Korea
- 2DNA Link, Inc., Seoul, Korea
- 3Department of Forensic Medicine, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2557779
- DOI: http://doi.org/10.3346/jkms.2024.39.e198
Abstract
- Background
Relatives share more genomic regions than unrelated individuals, with closer relatives sharing more regions. This concept, paired with the increased availability of highthroughput single nucleotide polymorphism (SNP) genotyping technologies, has made it feasible to measure the shared chromosomal regions between individuals to assess their level of relation to each other. However, such techniques have remained in the conceptual rather than practical stages in terms of applying measures or indices. Recently, we developed an index called “genetic distance-based index of chromosomal sharing (GD-ICS)” utilizing large-scale SNP data from Korean family samples and demonstrated its potential for practical applications in kinship determination. In the current study, we present validation results from various real cases demonstrating the utility of this method in resolving complex familial relationships where information obtained from traditional short tandem repeats (STRs) or lineage markers is inconclusive.
Methods
We obtained large-scale SNP data through microarray analysis from Korean individuals involving 13 kinship cases and calculated GD-ICS values using the method described in our previous study. Based on the GD-ICS reference constructed for Korean families, each disputed kinship was evaluated and validated using a combination of traditional STRs and lineage markers.
Results
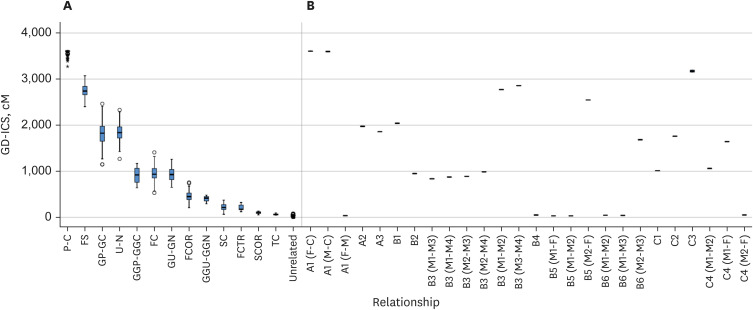
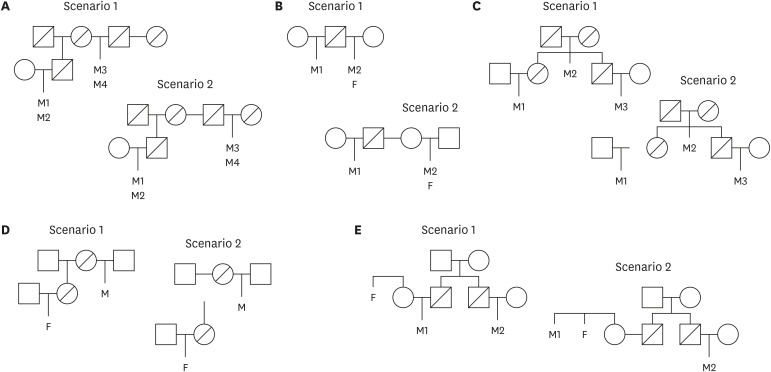
The cases comprised those A) that were found to be inconclusive using the traditional approach, B) for which it was difficult to apply traditional testing methods, and C) that were more conclusively resolved using the GD-ICS method. This method has overcome the limitations faced by traditional STRs in kinship testing, particularly in a paternity case with STR mutational events and in confirming distant kinship where the individual of interest is unavailable for testing. It has also been demonstrated to be effective in identifying various relationships without specific presumptions and in confirming a lack of genetic relatedness between individuals.
Conclusion
This method has been proven effective in identifying familial relationships across diverse complex and practical scenarios. It is not only useful when traditional testing methods fail to provide conclusive results, but it also enhances the resolution of challenging kinship cases, which suggests its applicability in various types of practical casework.
Keyword
Figure
Reference
-
1. Hill WG, White IMS. Identification of pedigree relationship from genome sharing. G3 (Gethesda). 2013; 3(9):1553–1571.2. Li R, Li H, Peng D, Hao B, Wang Z, Huang E, et al. Improved pairwise kinship analysis using massively parallel sequencing. Forensic Sci Int Genet. 2019; 38:77–85. PMID: 30368110.3. Thomson JA, Ayres KL, Pilotti V, Barrett MN, Walker JI, Debenham PG. Analysis of disputed single-parent/child and sibling relationships using 16 STR loci. Int J Legal Med. 2001; 115(3):128–134. PMID: 11775014.4. Pu CE, Linacre A. Increasing the confidence in half-sibship determination based upon 15 STR loci. J Forensic Leg Med. 2008; 15(6):373–377. PMID: 18586207.5. Sun M, Jobling MA, Taliun D, Pramstaller PP, Egeland T, Sheehan NA. On the use of dense SNP marker data for the identification of distant relative pairs. Theor Popul Biol. 2016; 107:14–25. PMID: 26474828.6. Kling D, Tillmar A. Forensic genealogy-A comparison of methods to infer distant relationships based on dense SNP data. Forensic Sci Int Genet. 2019; 42:113–124. PMID: 31302460.7. Morimoto C, Manabe S, Kawaguchi T, Kawai C, Fujimoto S, Hamano Y, et al. Pairwise kinship analysis by the index of chromosomal sharing using high-density single nucleotide polymorphisms. PLoS One. 2016; 11(7):e0160287. PMID: 27472558.8. Morimoto C, Manabe S, Fujimoto S, Hamano Y, Tamaki K. Discrimination of relationships with the same degree of kinship using chromosomal sharing patterns estimated from high-density SNPs. Forensic Sci Int Genet. 2018; 33:10–16. PMID: 29172066.9. Cho S, Shin E, Park YG, Choi SH, Choe EK, Bae JH, et al. A novel approach of kinship determination based on the physical length of genetically shared regions of chromosomes. Genes Genomics. 2024; 46(5):577–587. PMID: 38180716.10. Kim YL, Hwang JY, Kim YJ, Lee S, Chung NG, Goh HG, et al. Allele frequencies of 15 STR loci using AmpF/STR Identifiler kit in a Korean population. Forensic Sci Int. 2003; 136(1-3):92–95. PMID: 12969628.11. Hong SB, Kim SH, Kim KC, Park MH, Lee JY, Song JM, et al. Korean population genetic data and concordance for the PowerPlex® ESX 17, AmpFlSTR Identifiler®, and PowerPlex® 16 systems. Forensic Sci Int Genet. 2013; 7(3):e47–e51. PMID: 23419781.12. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81(3):559–575. PMID: 17701901.13. Delaneau O, Coulonges C, Zagury JF. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics. 2008; 9(1):540. PMID: 19087329.14. Jeong SJ, Lee JW, Lee SD, Lee SH, Park SJ, Kim JS, et al. Statistical evaluation of common relationships using STR markers in Korean population. J Sci Crim Invest. 2016; 10(2):107–115.15. Lindner I, von Wurmb-Schwark N, Meier P, Fimmers R, Büttner A. Usefulness of SNPs as supplementary markers in a paternity case with 3 genetic incompatibilities at autosomal and Y chromosomal loci. Transfus Med Hemother. 2014; 41(2):117–121. PMID: 24847187.16. Li L, Lin Y, Liu Y, Zhu R, Zhao Z, Que T. A case of false mother included with 46 autosomal STR markers. Investig Genet. 2015; 6(1):9.17. Tamura T, Osawa M, Ochiai E, Suzuki T, Nakamura T. Evaluation of advanced multiplex short tandem repeat systems in pairwise kinship analysis. Leg Med (Tokyo). 2015; 17(5):320–325. PMID: 25851967.18. Turrina S, Ferrian M, Caratti S, Cosentino E, De Leo D. Kinship analysis: assessment of related vs unrelated based on defined pedigrees. Int J Legal Med. 2016; 130(1):113–119. PMID: 26590134.19. Pinto N, Simões R, Amorim A, Conde-Sousa E. Optimizing the information increase through the addition of relatives and genetic markers in identification and kinship cases. Forensic Sci Int Genet. 2019; 40:210–218. PMID: 30921688.20. Snedecor J, Fennell T, Stadick S, Homer N, Antunes J, Stephens K, et al. Fast and accurate kinship estimation using sparse SNPs in relatively large database searches. Forensic Sci Int Genet. 2022; 61:102769. PMID: 36087514.21. de Vries JH, Kling D, Vidaki A, Arp P, Kalamara V, Verbiest MM, et al. Impact of SNP microarray analysis of compromised DNA on kinship classification success in the context of investigative genetic genealogy. Forensic Sci Int Genet. 2022; 56:102625. PMID: 34753062.22. Chu MC, Morimoto C, Kawai C, Miyao M, Tamaki K. Effects of DNA degradation and genotype imputation on high-density SNP microarray in pairwise kinship analysis. Leg Med (Tokyo). 2023; 60:102158. PMID: 36308842.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Analysis of Exophiala Species Using Molecular Markers
- Shared Decision Making in Geriatric Care
- Non-Synteny Regions in the Human Genome
- Inhaled Corticosteroids May Not Affect the Clinical Outcomes of Pneumonia in Patients with Chronic Obstructive Pulmonary Disease
- The Clinical Significance of Shared Epitope in Rheumatoid Arthritis