Korean J healthc assoc Infect Control Prev.
2024 Jun;29(1):19-26. 10.14192/kjicp.2024.29.1.19.
Laboratory Diagnosis and Interpretation of Urinary Tract Infections
- Affiliations
-
- 1Department of Laboratory Medicine, Kyung Hee University Hospital, Kyung Hee University College of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Korea University Guro Hospital, Korea University College of Medicine, Seoul, Korea
- KMID: 2556870
- DOI: http://doi.org/10.14192/kjicp.2024.29.1.19
Abstract
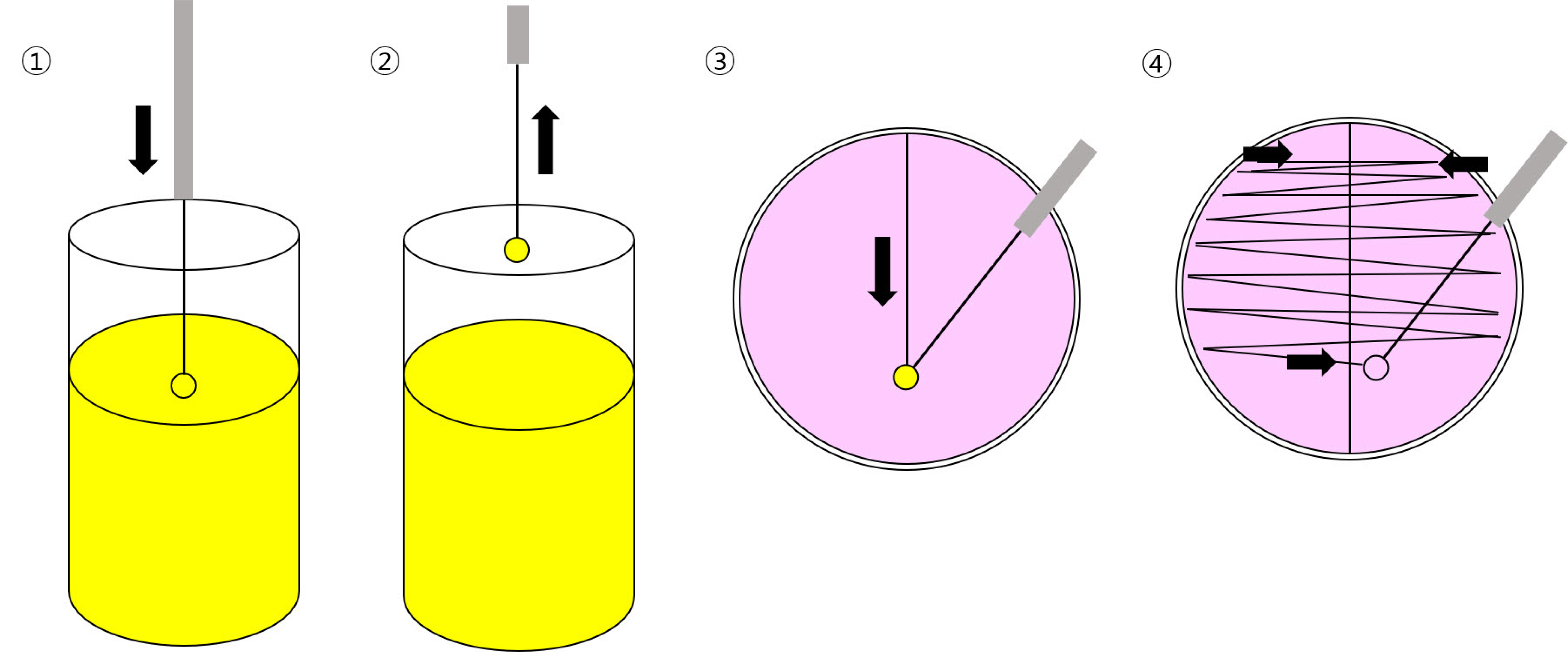
- Urinary tract infection (UTI) is a common and costly disease that affects millions of people worldwide each year. Accurate laboratory diagnosis of UTI is crucial to reduce antimicrobial resistance in UTI pathogens because of unnecessary antimicrobial use. Routine urinalysis with urine dipstick analysis and Gram staining can be used to screen for UTI. The conventional ‘gold standard’ for diagnosing UTIs involves culture-based tests. This method entails culturing the urine sample to amplify the bacteria to detectable levels, followed by biochemical and serological tests, as well as antimicrobial susceptibility tests. However, culture-based tests have the disadvantage of requiring 48-72 hours to report results owing to the time needed for bacterial growth. Therefore, various methods have been developed and are used to diagnose UTI to replace time-consuming culture tests. These methods include flow cytometry, mass spectrometry, and nucleic-acid-based diagnostic tests. This review introduces various laboratory methods used to diagnose UTI in clinical microbiology laboratories and discusses their principles and interpretation methods.
Keyword
Figure
Reference
-
1. Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015; Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 13:269–84. DOI: 10.1038/nrmicro3432. PMID: 25853778. PMCID: PMC4457377.2. Hrbacek J, Cermak P, Zachoval R. 2020; Current antibiotic resistance trends of uropathogens in Central Europe: survey from a tertiary hospital urology department 2011-2019. Antibiotics (Basel). 9:630. DOI: 10.3390/antibiotics9090630. PMID: 32971752. PMCID: PMC7559630.3. Chandra H, Singh C, Kumari P, Yadav S, Mishra AP, Laishevtcev A, et al. 2020; Promising roles of alternative medicine and plant-based nanotechnology as remedies for urinary tract infections. Molecules. 25:5593. DOI: 10.3390/molecules25235593. PMID: 33260701. PMCID: PMC7731396.4. Fritzenwanker M, Imirzalioglu C, Chakraborty T, Wagenlehner FM. 2016; Modern diagnostic methods for urinary tract infections. Expert Rev Anti Infect Ther. 14:1047–63. DOI: 10.1080/14787210.2016.1236685. PMID: 27624932.5. Ipe DS, Sundac L, Benjamin WH Jr, Moore KH, Ulett GC. 2013; Asymptomatic bacteriuria: prevalence rates of causal microorganisms, etiology of infection in different patient populations, and recent advances in molecular detection. FEMS Microbiol Lett. 346:1–10. DOI: 10.1111/1574-6968.12204. PMID: 23808987.6. Durkin MJ, Keller M, Butler AM, Kwon JH, Dubberke ER, Miller AC, et al. 2018; An assessment of inappropriate antibiotic use and guideline adherence for uncomplicated urinary tract infections. Open Forum Infect Dis. 5:ofy198. DOI: 10.1093/ofid/ofy198. PMID: 30191156. PMCID: PMC6121225.7. Fleming-Dutra KE, Hersh AL, Shapiro DJ, Bartoces M, Enns EA, File TM Jr, et al. 2016; Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010-2011. JAMA. 315:1864–73. DOI: 10.1001/jama.2016.4151. PMID: 27139059.8. Sanchez GV, Master RN, Karlowsky JA, Bordon JM. 2012; In vitro antimicrobial resistance of urinary Escherichia coli isolates among U.S. outpatients from 2000 to 2010. Antimicrob Agents Chemother. 56:2181–3. DOI: 10.1128/AAC.06060-11. PMID: 22252813. PMCID: PMC3318377.9. Franco-Duarte R, Černáková L, Kadam S, Kaushik KS, Salehi B, Bevilacqua A, et al. 2019; Advances in chemical and biological methods to identify microorganisms-from past to present. Microorganisms. 7:130. DOI: 10.3390/microorganisms7050130. PMID: 31086084. PMCID: PMC6560418.10. Davenport M, Mach KE, Shortliffe LMD, Banaei N, Wang TH, Liao JC. 2017; New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol. 14:296–310. DOI: 10.1038/nrurol.2017.20. PMID: 28248946. PMCID: PMC5473291.11. Korean Society for Laboratory Medicine. 2021. Laboratory medicine. 6th ed. Panmun Education;Seoul: p. 687–9.12. Chong Y, Lee K, Kim HS, Shin JH, Jeong SH, Yong D, et al. 2022. Diagnostic microbiology. 7th ed. Seo Heung Publishing Company;Seoul: p. 114–7.13. Carroll KC, Pfaller MA, Landry ML, McAdam AJ, Patel R, Richter SS, et al. 2019. Manual of clinical microbiology. 12th ed. ASM Press;Washington, DC: p. 302–30. DOI: 10.1128/9781555819842.14. Eisinger SW, Schwartz M, Dam L, Riedel S. 2013; Evaluation of the BD Vacutainer Plus Urine C&S Preservative Tubes compared with nonpreservative urine samples stored at 4°C and room temperature. Am J Clin Pathol. 140:306–13. DOI: 10.1309/AJCP5ON9JHXVNQOD. PMID: 23955448.15. McTaggart SJ. 2005; Childhood urinary conditions. Aust Fam Physician. 34:937–41.16. Patel HP. 2006; The abnormal urinalysis. Pediatr Clin North Am. 53:325–37. v. DOI: 10.1016/j.pcl.2006.02.004. PMID: 16716783.17. Orellana MA, Gómez-Lus ML, Lora D. 2012; Sensitivity of Gram stain in the diagnosis of urethritis in men. Sex Transm Infect. 88:284–7. DOI: 10.1136/sextrans-2011-050150. PMID: 22308534.18. Karah N, Rafei R, Elamin W, Ghazy A, Abbara A, Hamze M, et al. 2020; Guideline for urine culture and biochemical identification of bacterial urinary pathogens in low-resource settings. Diagnostics (Basel). 10:832. DOI: 10.3390/diagnostics10100832. PMID: 33081114. PMCID: PMC7602787.19. Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, et al. 2016; The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 54:1216–22. DOI: 10.1128/JCM.00044-16. PMID: 26962083. PMCID: PMC4844725.20. Deen NS, Ahmed A, Tasnim NT, Khan N. 2023; Clinical relevance of expanded quantitative urine culture in health and disease. Front Cell Infect Microbiol. 13:1210161. DOI: 10.3389/fcimb.2023.1210161. PMID: 37593764. PMCID: PMC10428011.21. Thapaliya J, Khadka P, Thapa S, Gongal C. 2020; Enhanced quantitative urine culture technique, a slight modification, in detecting under-diagnosed pediatric urinary tract infection. BMC Res Notes. 13:5. DOI: 10.1186/s13104-019-4875-y. PMID: 31900212. PMCID: PMC6942300.22. Lewis II JS, Mathers AJ, Bobenchik AM, Bryson AL, Campeau S, Cullen SK, et al. Clinical and Laboratory Standards Institute (CLSI). 2024. CLSI M100: performance standards for antimicrobial susceptibility testing. 34th ed. CLSI;Berwyn:23. Previtali G, Ravasio R, Seghezzi M, Buoro S, Alessio MG. 2017; Performance evaluation of the new fully automated urine particle analyser UF-5000 compared to the reference method of the Fuchs-Rosenthal chamber. Clin Chim Acta. 472:123–30. DOI: 10.1016/j.cca.2017.07.028. PMID: 28760666.24. Wang H, Han FF, Wen JX, Yan Z, Han YQ, Hu ZD, et al. 2023; Accuracy of the Sysmex UF-5000 analyzer for urinary tract infection screening and pathogen classification. PLoS One. 18:e0281118. DOI: 10.1371/journal.pone.0281118. PMID: 36724192. PMCID: PMC9891513.25. Enko D, Stelzer I, Böckl M, Schnedl WJ, Meinitzer A, Herrmann M, et al. 2021; Comparison of the reliability of Gram-negative and Gram-positive flags of the Sysmex UF-5000 with manual Gram stain and urine culture results. Clin Chem Lab Med. 59:619–24. DOI: 10.1515/cclm-2020-1263. PMID: 33068381.26. Ferreira L, Sánchez-Juanes F, García-Fraile P, Rivas R, Mateos PF, Martínez-Molina E, et al. 2011; MALDI-TOF mass spectrometry is a fast and reliable platform for identification and ecological studies of species from family Rhizobiaceae. PLoS One. 6:e20223. DOI: 10.1371/journal.pone.0020223. PMID: 21655291. PMCID: PMC3105015.27. Demarco ML, Burnham CA. 2014; Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. Am J Clin Pathol. 141:204–12. DOI: 10.1309/AJCPQYW3B6JLKILC. PMID: 24436267.28. Burillo A, Rodríguez-Sánchez B, Ramiro A, Cercenado E, Rodríguez-Créixems M, Bouza E. 2014; Gram-stain plus MALDI-TOF MS (matrix-assisted laser desorption ionization-time of flight mass spectrometry) for a rapid diagnosis of urinary tract infection. PLoS One. 9:e86915. DOI: 10.1371/journal.pone.0086915. PMID: 24466289. PMCID: PMC3899310.29. Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007; A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 69:330–9. DOI: 10.1016/j.mimet.2007.02.005. PMID: 17391789. PMCID: PMC2562909.30. Leber AL, Burnham CAD. 2023. Clinical microbiology procedures handbook. 5th ed. ASM Press;Washington, DC: p. 1–12.31. Imirzalioglu C, Hain T, Chakraborty T, Domann E. 2008; Hidden pathogens uncovered: metagenomic analysis of urinary tract infections. Andrologia. 40:66–71. DOI: 10.1111/j.1439-0272.2007.00830.x. PMID: 18336452.32. Aguilera-Arreola MG, Martínez-Peña MD, Hernández-Martínez F, Juárez Enriques SR, Rico Verdín B, Majalca-Martínez C, et al. 2016; Cultivation-independent approach for the direct detection of bacteria in human clinical specimens as a tool for analysing culture-negative samples: a prospective study. Springerplus. 5:332. DOI: 10.1186/s40064-016-1949-3. PMID: 27065040. PMCID: PMC4792836.33. Domann E, Hong G, Imirzalioglu C, Turschner S, Kühle J, Watzel C, et al. 2003; Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients. J Clin Microbiol. 41:5500–10. DOI: 10.1128/JCM.41.12.5500-5510.2003. PMID: 14662931. PMCID: PMC309025.34. van der Zee A, Roorda L, Bosman G, Ossewaarde JM. 2016; Molecular diagnosis of urinary tract infections by semi-quantitative detection of uropathogens in a routine clinical hospital setting. PLoS One. 11:e0150755. DOI: 10.1371/journal.pone.0150755. PMID: 26954694. PMCID: PMC4783162.35. Wojno KJ, Baunoch D, Luke N, Opel M, Korman H, Kelly C, et al. 2020; Multiplex PCR based urinary tract infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology. 136:119–26. DOI: 10.1016/j.urology.2019.10.018. PMID: 31715272.36. Perez-Carrasco V, Soriano-Lerma A, Soriano M, Gutiérrez-Fernández J, Garcia-Salcedo JA. 2021; Urinary microbiome: yin and yang of the urinary tract. Front Cell Infect Microbiol. 11:617002. DOI: 10.3389/fcimb.2021.617002. PMID: 34084752. PMCID: PMC8167034.37. Moustafa A, Li W, Singh H, Moncera KJ, Torralba MG, Yu Y, et al. 2018; Microbial metagenome of urinary tract infection. Sci Rep. 8:4333. DOI: 10.1038/s41598-018-22660-8. PMID: 29531289. PMCID: PMC5847550.38. Szlachta-McGinn A, Douglass KM, Chung UYR, Jackson NJ, Nickel JC, Ackerman AL. 2022; Molecular diagnostic methods versus conventional urine culture for diagnosis and treatment of urinary tract infection: a systematic review and meta-analysis. Eur Urol Open Sci. 44:113–24. DOI: 10.1016/j.euros.2022.08.009. PMID: 36093322. PMCID: PMC9459428.39. Almas S, Carpenter RE, Rowan C, Tamrakar VK, Bishop J, Sharma R. 2023; Advantage of precision metagenomics for urinary tract infection diagnostics. Front Cell Infect Microbiol. 13:1221289. DOI: 10.3389/fcimb.2023.1221289. PMID: 37469596. PMCID: PMC10352793.40. Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, Frimodt-Møller N, et al. 2014; Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol. 52:139–46. DOI: 10.1128/JCM.02452-13. PMID: 24172157. PMCID: PMC3911411.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Guideline for the Diagnosis and Treatment of Urinary Tract Infections: Asymptomatic Bacteriuria, Uncomplicated & Complicated Urinary Tract Infections, Bacterial Prostatitis
- Urinary Tract Infection and Its Diagnosis in Children
- Diagnosis and treatment of urinary tract infection in adults
- Treatment of urinary tract infections
- Diagnosis of urinary tract infection in children