Cancer Res Treat.
2024 Apr;56(2):665-674. 10.4143/crt.2023.864.
Case Series of Soft Tissue Sarcoma Patients with Brain Metastasis with Implications from Genomic and Transcriptomic Analysis
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 2Department of Biomedical Sciences, Seoul National University College of Medicine, Seoul, Korea
- 3Genomic Medicine Institute, Medical Research Center, Seoul National University, Seoul, Korea
- 4Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea
- 5Department of Orthopaedic Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2554355
- DOI: http://doi.org/10.4143/crt.2023.864
Abstract
- Purpose
Brain metastasis rarely occurs in soft tissue sarcoma (STS). Here, we present five cases of STS with brain metastases with genetic profiles.
Materials and Methods
We included five patients from Seoul National University Hospital who were diagnosed with STS with metastasis to the brain. Tissue from the brain metastasis along with that from the primary site or other metastases were used for DNA and RNA sequencing to identify genetic profiles. Gene expression profiles were compared with sarcoma samples from The Cancer Genome Atlas.
Results
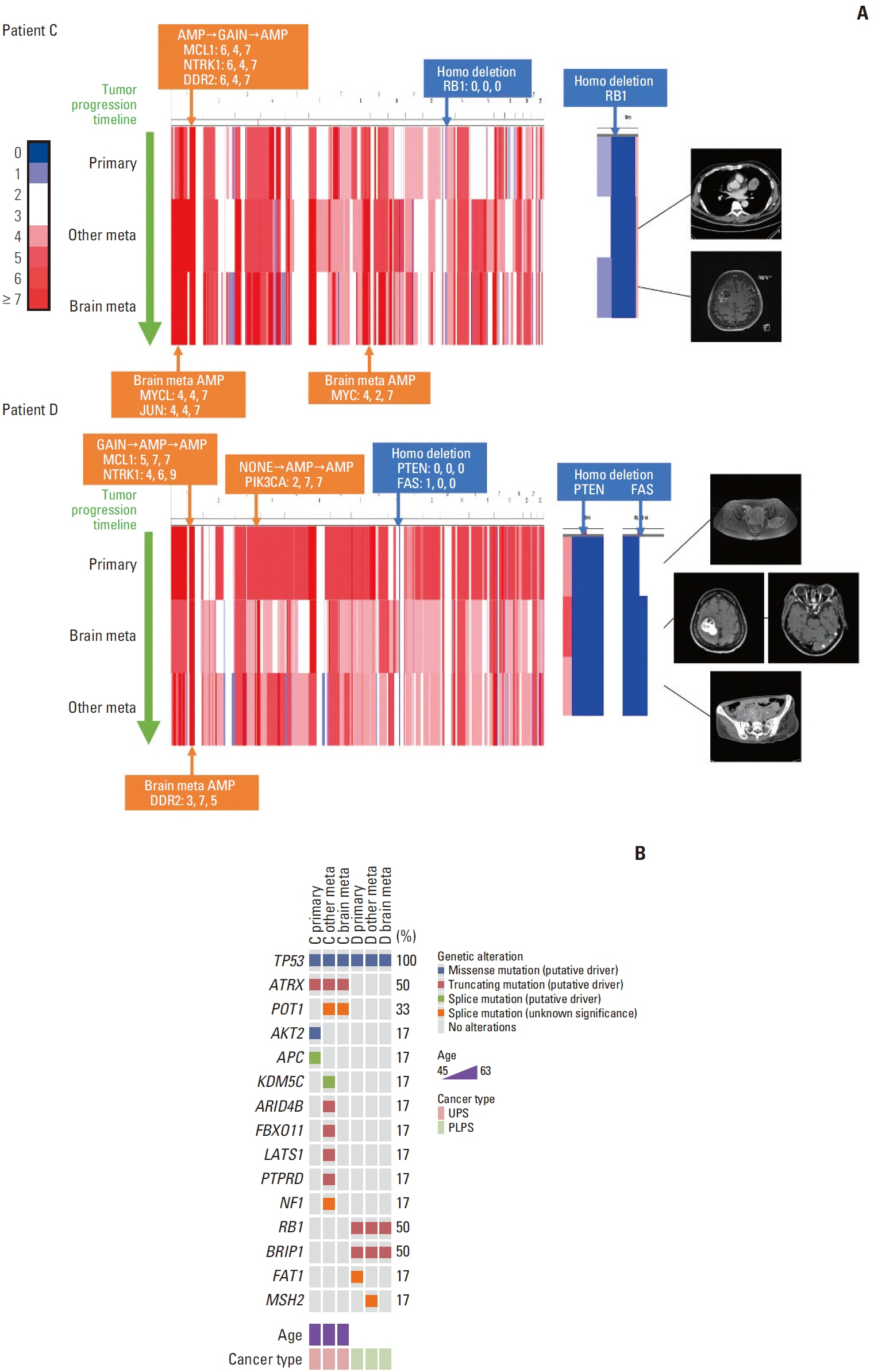
The overall survival after diagnosis of brain metastasis ranged from 2.2 to 34.3 months. Comparison of mutational profiles between brain metastases and matched primary or other metastatic samples showed similar profiles. In two patients, copy number variation profiles between brain metastasis and other tumors showed several differences including MYCL, JUN, MYC, and DDR2 amplification. Gene ontology analysis showed that the group of genes significantly highly expressed in the brain metastasis samples was enriched in the G-protein coupled receptor activity, structural constituent of chromatin, protein heterodimerization activity, and binding of DNA, RNA, and protein. Gene set enrichment analysis showed enrichment in the pathway of neuroactive ligand-receptor interaction and systemic lupus erythematosus.
Conclusion
The five patients had variable ranges of clinical courses and outcomes. Genomic and transcriptomic analysis of STS with brain metastasis implicates possible involvement of complex expression modification and epigenetic changes rather than the addition of single driver gene alteration.
Keyword
Figure
Reference
-
References
1. Cancer Genome Atlas Research Network. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017; 171:950–96.2. Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013; 5:147–62.3. Al Sannaa G, Watson KL, Olar A, Wang WL, Fuller GN, McCutcheon I, et al. Sarcoma brain metastases: 28 years of experience at a single institution. Ann Surg Oncol. 2016; 23:962–7.
Article4. Chaigneau L, Patrikidou A, Ray-Coquard I, Valentin T, Linassier C, Bay JO, et al. Brain metastases from adult sarcoma: prognostic factors and impact of treatment. A retrospective analysis from the French Sarcoma Group (GSF/GETO). Oncologist. 2018; 23:948–55.
Article5. Shweikeh F, Bukavina L, Saeed K, Sarkis R, Suneja A, Sweiss F, et al. Brain metastasis in bone and soft tissue cancers: a review of incidence, interventions, and outcomes. Sarcoma. 2014; 2014:475175.
Article6. Salvati M, D’Elia A, Frati A, Santoro A. Sarcoma metastatic to the brain: a series of 35 cases and considerations from 27 years of experience. J Neurooncol. 2010; 98:373–7.
Article7. Gercovich FG, Luna MA, Gottlieb JA. Increased incidence of cerebral metastases in sarcoma patients with prolonged survival from chemotherapy: report of cases of leiomysarcoma and chondrosarcoma. Cancer. 1975; 36:1843–51.
Article8. Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015; 5:1164–77.9. Fischer GM, Jalali A, Kircher DA, Lee WC, McQuade JL, Haydu LE, et al. Molecular profiling reveals unique immune and metabolic features of melanoma brain metastases. Cancer Discov. 2019; 9:628–45.
Article10. Vareslija D, Priedigkeit N, Fagan A, Purcell S, Cosgrove N, O’Halloran PJ, et al. Transcriptome characterization of matched primary breast and brain metastatic tumors to detect novel actionable targets. J Natl Cancer Inst. 2019; 111:388–98.
Article11. Shih DJ, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet. 2020; 52:371–7.
Article12. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017:PO.17.00011.
Article13. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011; 29:24–6.
Article14. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4.
Article15. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6:pl1.
Article16. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25.
Article17. Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020; 38:675–8.
Article18. Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022; 50:W216–21.19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–50.
Article20. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30.
Article21. Nacev BA, Sanchez-Vega F, Smith SA, Antonescu CR, Rosenbaum E, Shi H, et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022; 13:3405.
Article22. Demicco EG. Molecular updates in adipocytic neoplasms. Semin Diagn Pathol. 2019; 36:85–94.
Article23. Jaber OI, Kirby PA. Alveolar soft part sarcoma. Arch Pathol Lab Med. 2015; 139:1459–62.
Article24. Chen G, Chakravarti N, Aardalen K, Lazar AJ, Tetzlaff MT, Wubbenhorst B, et al. Molecular profiling of patient-matched brain and extracranial melanoma metastases implicates the PI3K pathway as a therapeutic target. Clin Cancer Res. 2014; 20:5537–46.
Article25. Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022; 185:563–75.26. Hofvander J, Viklund B, Isaksson A, Brosjo O, Vult von Steyern F, Rissler P, et al. Different patterns of clonal evolution among different sarcoma subtypes followed for up to 25 years. Nat Commun. 2018; 9:3662.
Article27. Xie W, Zhang J, Zhong P, Qin S, Zhang H, Fan X, et al. Expression and potential prognostic value of histone family gene signature in breast cancer. Exp Ther Med. 2019; 18:4893–903.
Article28. Zhang L, Fan M, Napolitano F, Gao X, Xu Y, Li L. Transcriptomic analysis identifies organ-specific metastasis genes and pathways across different primary sites. J Transl Med. 2021; 19:31.
Article29. Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011; 11:597–606.
Article30. Chibon F, Lagarde P, Salas S, Perot G, Brouste V, Tirode F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010; 16:781–7.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Alveolar soft part Sarcoma with Metastasis to Bone: A Case Report
- Intracerebral Metastasis of Alveolar Soft Part Sarcoma: A case report and study on its histogenesis
- A Case of Intracerebral Metastasis of Alveolar Soft Part Sarcoma: Case Report
- Cerebral Metastases and Menifestation of the Alveolar Soft Part Sarcoma
- Primary pulmonary synovial sarcoma with brain metastasis