Ewha Med J.
2024 Jan;47(1):e6. 10.12771/emj.2024.e6.
Exosomal microRNAs (miRNAs) in blood and urine under physiological conditions: a comparative study
- Affiliations
-
- 1Basic Medical College, Chengdu University, Chengdu, China
- 2Nephrology Department, Affiliated Hospital of Chengdu University, Chengdu, China
- KMID: 2553298
- DOI: http://doi.org/10.12771/emj.2024.e6
Abstract
Objectives
Blood and urine are commonly used specimens for clinical testing, and their contents, particularly exosomal microRNA (miRNA), are diverse, reflecting the metabolic activities of tissues and organs in the body.
Methods
Blood and urine samples were collected from six healthy adults. Exosomes were then enriched from these samples, followed by sequencing and bioinformatic analysis of exosomal miRNA.
Results
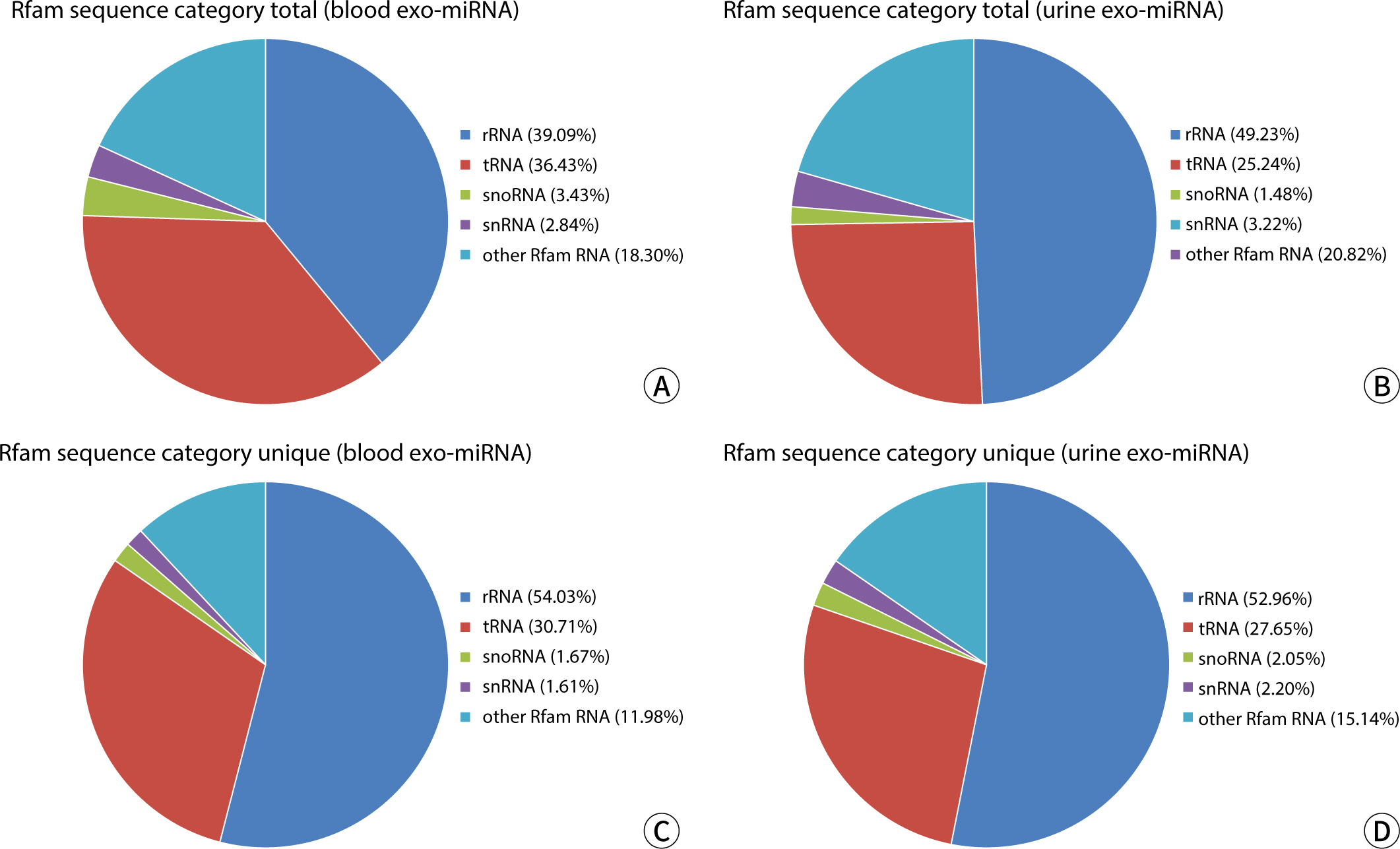
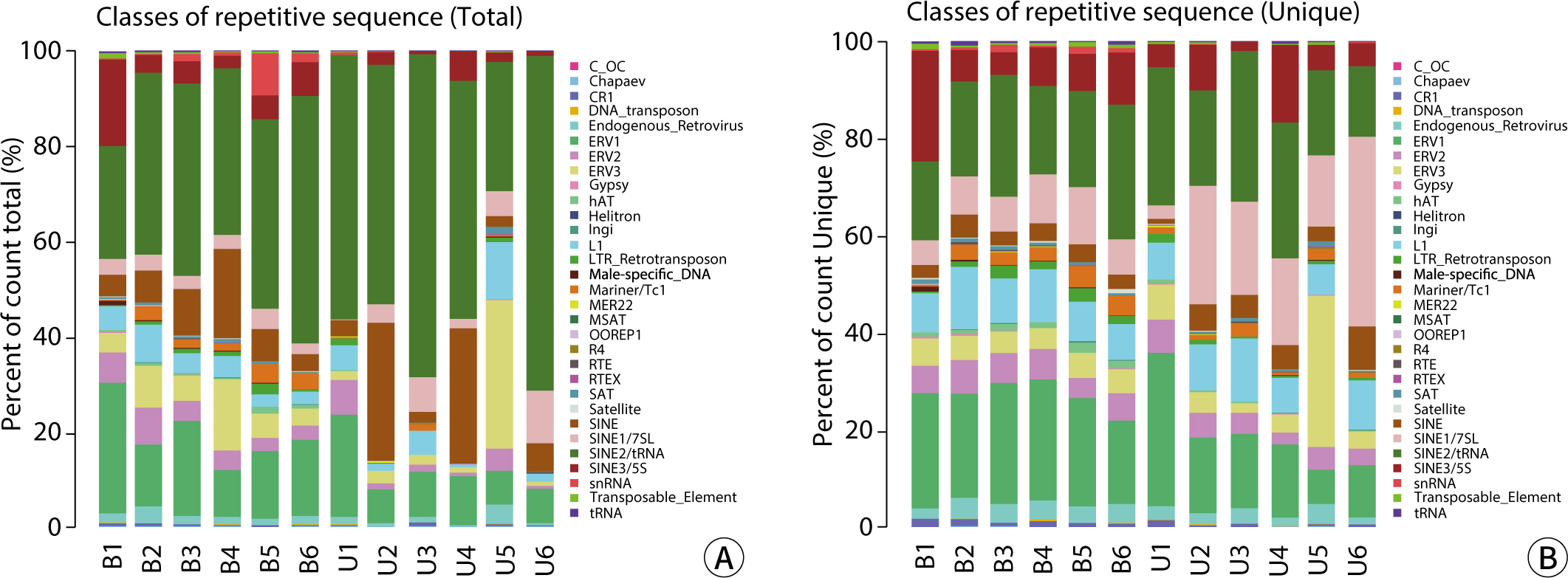
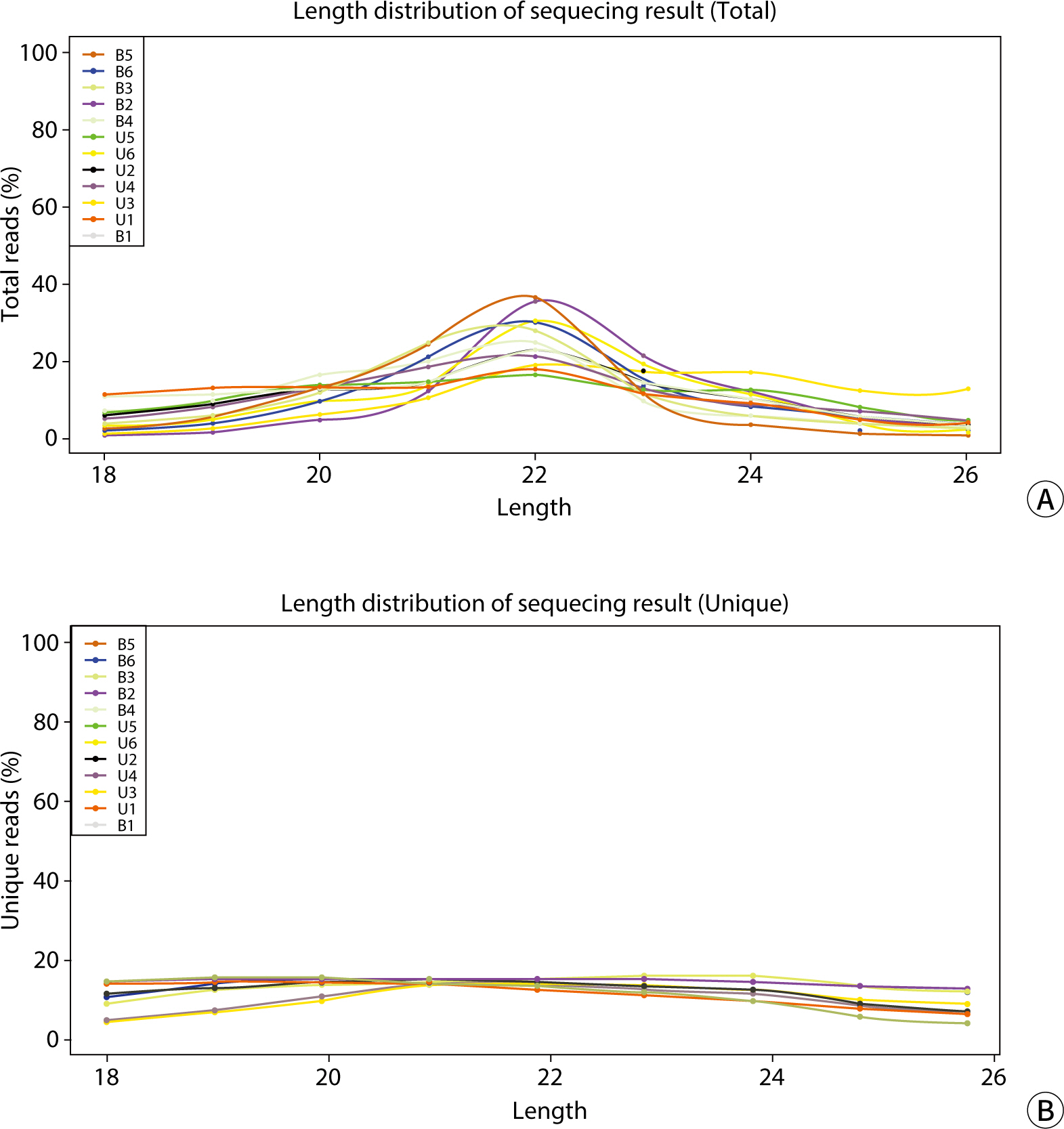
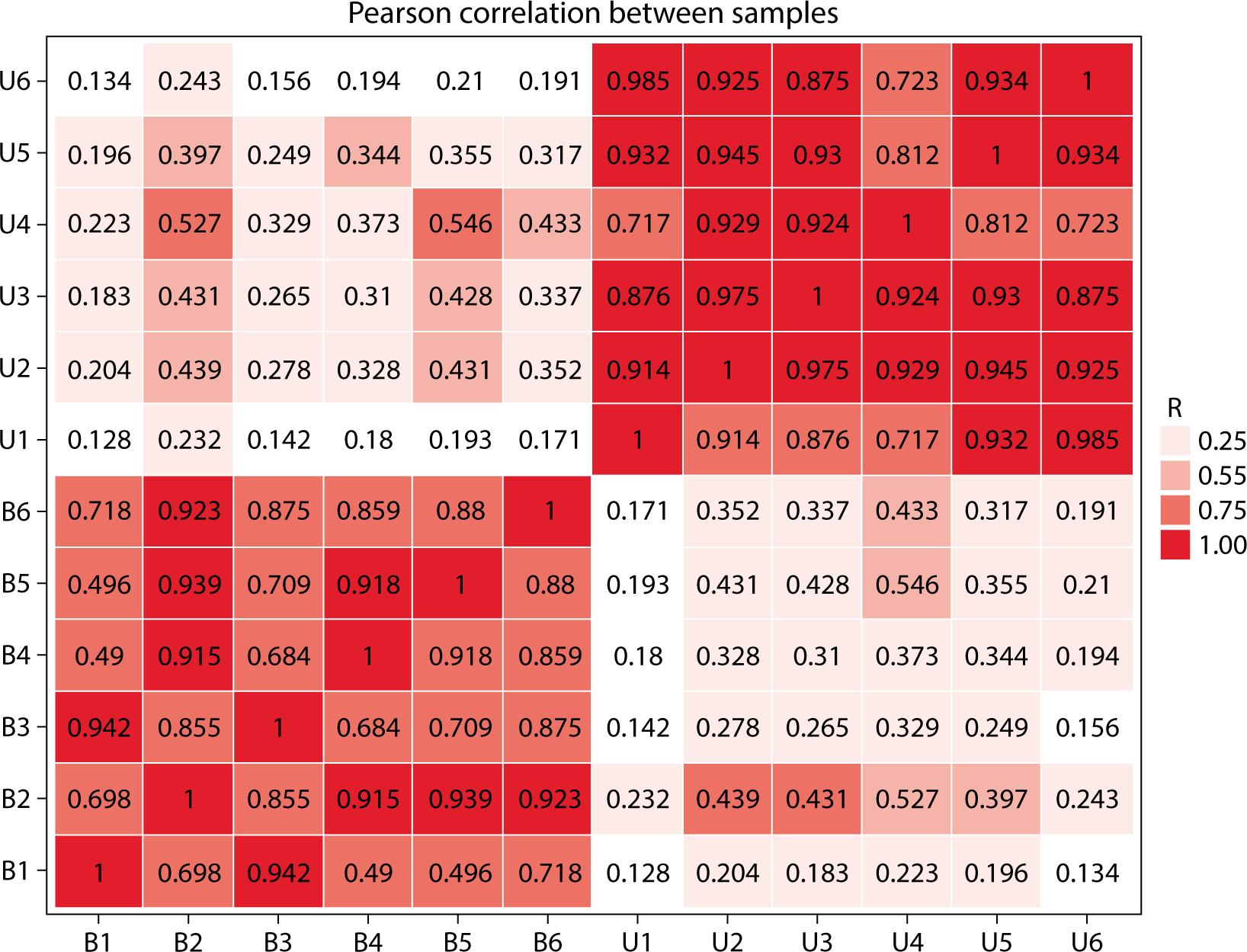
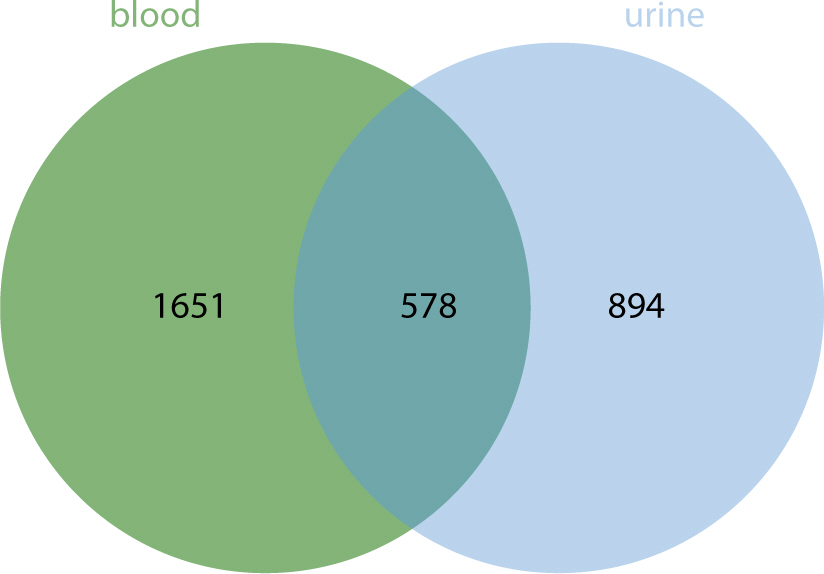
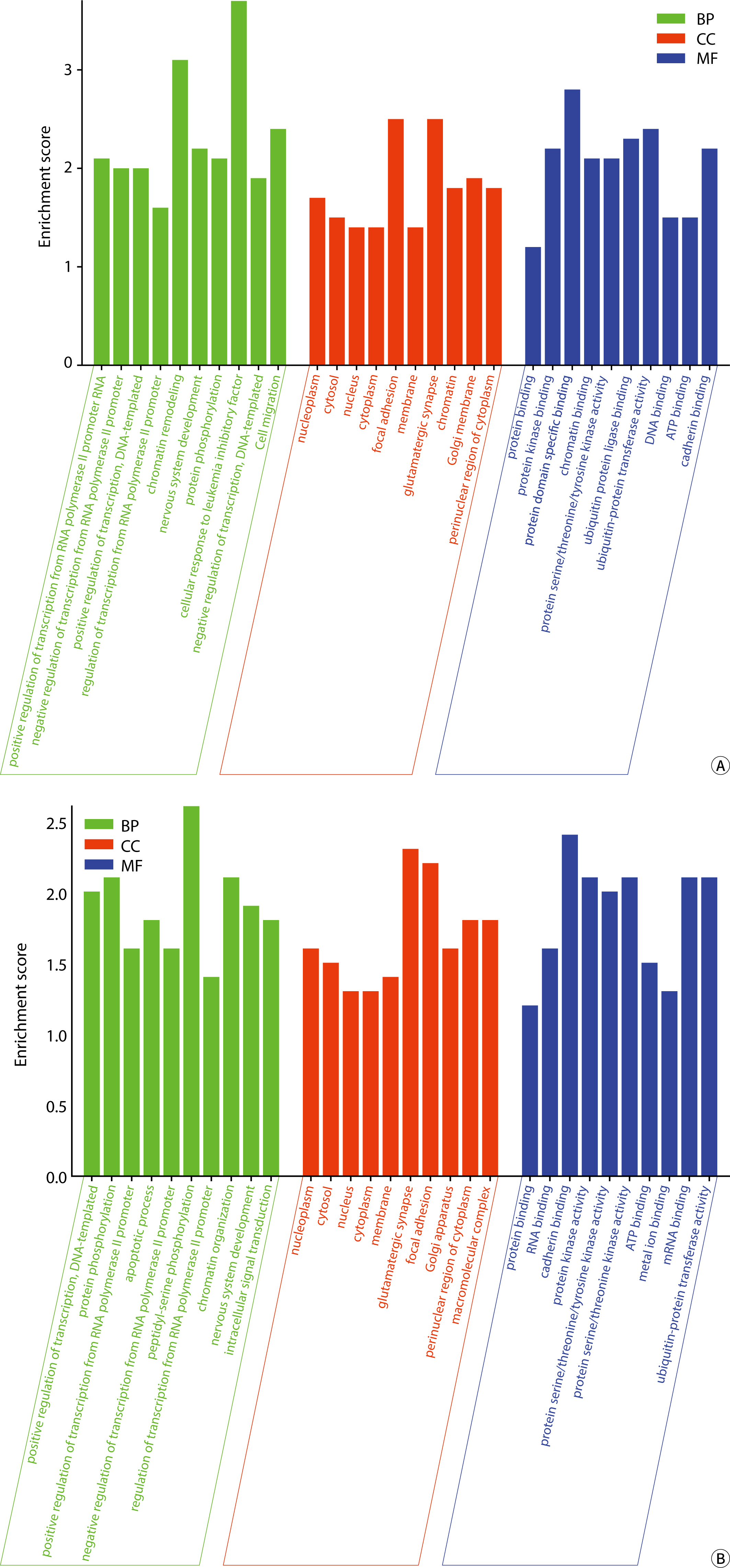
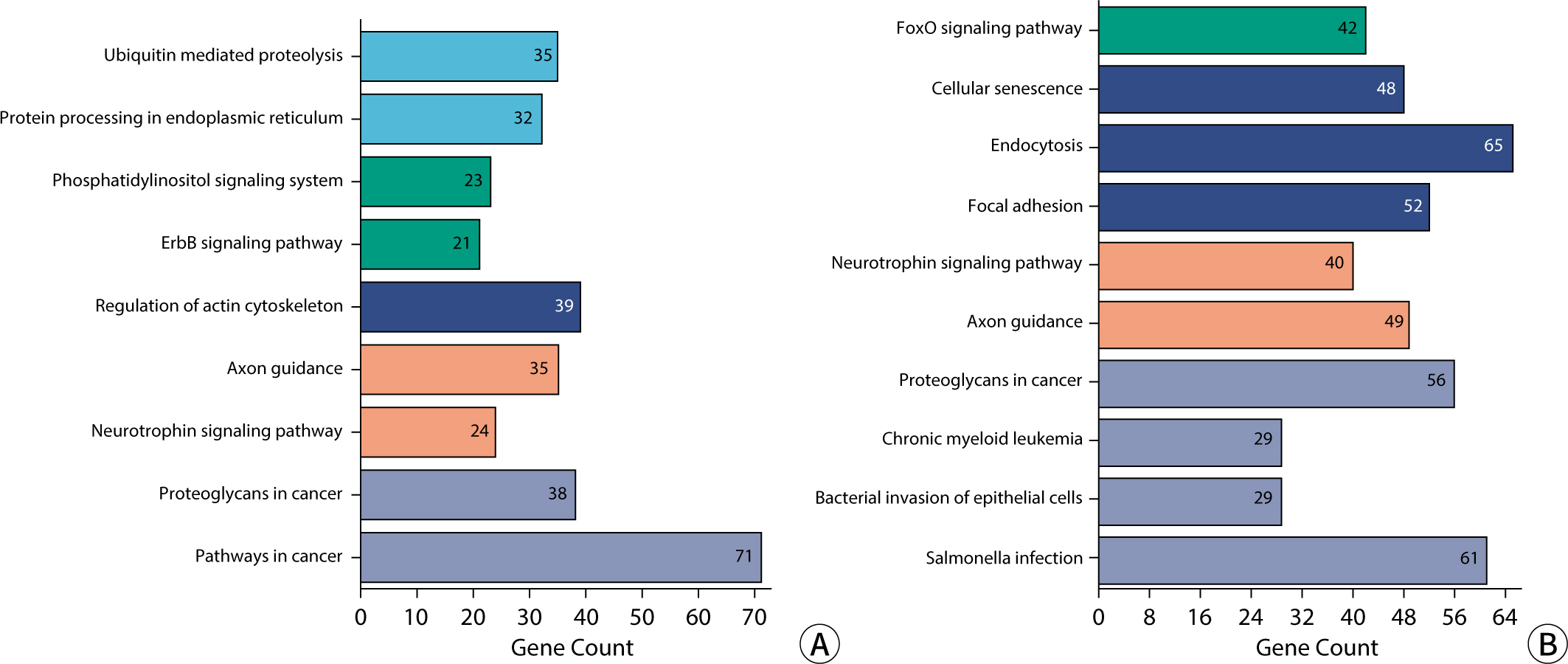
The comparative analysis of miRNAs in blood and urine revealed that 41 miRNAs were more abundant in blood, while 61 were found at lower levels. Notably, hsa-miR-934 was among those with higher expression in blood, whereas hsa-miR-425-5p was one of the miRNAs with lower expression. Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that the target mRNAs of differentially expressed exosomal miRNAs (DEexo-miRNAs) in both blood and urine are implicated in various signaling pathways, including proteoglycans in cancer, axonal guidance, and the regulation of the actin cytoskeleton. Additionally, the target mRNAs associated with DEexo-miRNAs in urine were also linked to processes such as ubiquitin-mediated proteolysis and the phosphatidylinositol signaling system. In contrast, the target mRNAs corresponding to DEexo-miRNAs in blood were involved in the FoxO signaling pathway and chronic myeloid leukemia, among others.
Conclusion
This study observed differential expression of exosomal miRNAs in blood and urine, thereby enriching the available library of exosomal miRNA for these two sample types. It also lays the groundwork for the detection of exosomal biomarkers from blood and urine.
Keyword
Figure
Cited by 1 articles
-
Gender equity in medical journals in Korea and this issue
Sun Huh
Ewha Med J. 2024;47(1):e1. doi: 10.12771/emj.2024.e1.
Reference
-
References
1. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010; 79:351–379. DOI: 10.1146/annurev-biochem-060308-103103. PMID: 20533884.
Article2. Gupta J, Tayyib NA, Jalil AT, Hlail SH, Zabibah RS, Vokhidov UN, et al. Angiogenesis and prostate cancer: microRNAs comes into view. Pathol Res Pract. 2023; 248:154591. DOI: 10.1016/j.prp.2023.154591. PMID: 37343381.
Article3. Khordadmehr M, Matin R, Baradaran B, Baghbani E, Jigari-Asl F, Noorolyai S. The effect of miR-4800 restoration on proliferation and migration of human breast cancer cells in vitro. Adv Pharm Bull. 2023; 13(2):378–384. DOI: 10.34172/apb.2023.041. PMID: 37342379. PMCID: PMC10278211.
Article4. García-Sánchez D, González-González A, Alfonso-Fernández A, Del Dujo-Gutiérrez M, Pérez-Campo FM. Communication between bone marrow mesenchymal stem cells and multiple myeloma cells: impact on disease progression. World J Stem Cells. 2023; 15(5):421–437. DOI: 10.4252/wjsc.v15.i5.421. PMID: 37342223. PMCID: PMC10277973.
Article5. Yuan Y, Rao LZ, Zhang SH, Xu Y, Li TT, Zou J, et al. Exercise regulates bone metabolism via microRNAs. Acta Physiol Sin. 2023; 75(3):429–438.6. de Matos BM, Stimamiglio MA, Correa A, Robert AW. Human pluripotent stem cell-derived extracellular vesicles: from now to the future. World J Stem Cells. 2023; 15(5):453–465. DOI: 10.4252/wjsc.v15.i5.453. PMID: 37342215. PMCID: PMC10277970.
Article7. Yuan YG, Wang JL, Zhang YX, Li L, Reza AMMT, Gurunathan S. Biogenesis, composition and potential therapeutic applications of mesenchymal stem cells derived exosomes in various diseases. Int J Nanomed. 2023; 18:3177–3210. DOI: 10.2147/IJN.S407029. PMID: 37337578. PMCID: PMC10276992.
Article8. Jiang X, Zhang Z, Hou M, Yang X, Cui L. Plasma exosomes and contained MiRNAs affect the reproductive phenotype in polycystic ovary syndrome. FASEB J. 2023; 37(7):e22960. DOI: 10.1096/fj.202201940RR. PMID: 37335566.
Article9. Gu F, Jiang J, Sun P. Recent advances of exosomes in age-related macular degeneration. Front Pharmacol. 2023; 14:1204351. DOI: 10.3389/fphar.2023.1204351. PMID: 37332352. PMCID: PMC10272348.
Article10. Poulet G, Massias J, Taly V. Liquid biopsy: general concepts. Acta Cytol. 2019; 63(6):449–455. DOI: 10.1159/000499337. PMID: 31091522.
Article11. Lv C, Xu Y, Wang Y, Ding W, Yao D, Zhao Z, et al. Comparative analysis of urinary sediments and extracellular RNA in patients with renal fibrosis. Guangdong Med. 2020; 41(15):1567–1572.12. Lv C, Wang Y, Ding W. Effective isolation of urinary exosomes by novel approach: polyethylene glycol precipitation. Chin J Biochem Mol Biol. 2018; 34(1):110–116.13. Lv C, Ding W, Wang Y, Zhao Z, Li J, Chen Y, et al. A PEG-based method for the isolation of urinary exosomes and its application in renal fibrosis diagnostics using cargo miR-29c and miR-21 analysis. Int Urol Nephrol. 2018; 50(5):973–982. DOI: 10.1007/s11255-017-1779-4. PMID: 29330775.
Article14. Yu C, Zhang M, Xiong Y, Wang Q, Wang Y, Wu S, et al. Comparison of miRNa transcriptome of exosomes in three categories of somatic cells with derived iPSCs. Sci Data. 2023; 10(1):616. DOI: 10.1038/s41597-023-02493-5. PMID: 37696871. PMCID: PMC10495316.
Article15. Perez-Hernandez J, Riffo-Campos AL, Ortega A, Martinez-Arroyo O, Perez-Gil D, Olivares D, et al. Urinary- and plasma-derived exosomes reveal a distinct microRNA signature associated with albuminuria in hypertension. Hypertension. 2021; 77(3):960–971. DOI: 10.1161/HYPERTENSIONAHA.120.16598. PMID: 33486986.
Article16. Assmann T, Recamonde-Mendoza M, De Souza BM, Bauer AC, Crispim D. MicroRNAs and diabetic kidney disease: systematic review and bioinformatic analysis. Mol Cell Endocrinol. 2018; 477:90–102. DOI: 10.1016/j.mce.2018.06.005. PMID: 29902497.
Article17. Ma J, Zhu M, Ye X, Wu B, Wang T, Ma M, et al. Prognostic microRNAs associated with phosphoserine aminotransferase 1 in gastric cancer as markers of bone metastasis. Front Genet. 2022; 13:959684. DOI: 10.3389/fgene.2022.959684. PMID: 36061202. PMCID: PMC9437321.
Article18. Frørup C, Mirza AH, Yarani R, Nielsen LB, Mathiesen ER, Damm P, et al. Plasma exosome-enriched extracellular vesicles from lactating mothers with type 1 diabetes contain aberrant levels of mirNAS during the postpartum period. Front Immunol. 2021; 12:744509. DOI: 10.3389/fimmu.2021.744509. PMID: 34691048. PMCID: PMC8531745.19. Wu J, Feng Z, Wang R, Li A, Wang H, He X, et al. Integration of bioinformatics analysis and experimental validation identifies plasma exosomal miR-103b/877-5p/29c-5p as diagnostic biomarkers for early lung adenocarcinoma. Cancer Med. 2022; 11(23):4411–4421. DOI: 10.1002/cam4.4788. PMID: 35585716. PMCID: PMC9741994.
Article20. Chen D, Zhang G, Li Y, Zhang M, He Q, Yang J, et al. Up-regulation of urinary exosomal hsa-microRNA-200b-3p and hsa-microRNA-206 in patients of steroid-induced osteonecrosis of femoral head. Am J Transl Res. 2021; 13(7):7574–7590. DOI: 10.21203/rs.3.rs-68462/v1.
Article21. Sinha N, Puri V, Kumar V, Nada R, Rastogi A, Jha V, et al. Urinary exosomal miRNA-663a shows variable expression in diabetic kidney disease patients with or without proteinuria. Sci Rep. 2023; 13(1):4516. DOI: 10.1038/s41598-022-26558-4. PMID: 36934129. PMCID: PMC10024703.
Article22. Deng J, Li YQ, Liu Y, Li Q, Hu Y, Xu JQ, et al. Exosomes derived from plasma of septic patients inhibit apoptosis of T lymphocytes by down-regulating bad via hsa-miR-7-5p. Biochem Biophys Res Commun. 2019; 513(4):958–966. DOI: 10.1016/j.bbrc.2019.04.051. PMID: 31003766.
Article23. Zhang F, Li S, Zhang T, Yu B, Zhang J, Ding H, et al. High throughput microRNAs sequencing profile of serum exosomes in women with and without polycystic ovarian syndrome. PeerJ. 2021; 9:e10998. DOI: 10.7717/peerj.10998. PMID: 33763302. PMCID: PMC7958896.
Article24. Hang W, Feng Y, Sang Z, Yang Y, Zhu Y, Huang Q, et al. Downregulation of miR-145-5p in cancer cells and their derived exosomes may contribute to the development of ovarian cancer by targeting CT. Int J Mol Med. 2019; 43(1):256–266. DOI: 10.3892/ijmm.2018.3958.25. Zhao Z, Shuang T, Gao Y, Lu F, Zhang J, He W, et al. Targeted delivery of exosomal miR-484 reprograms tumor vasculature for chemotherapy sensitization. Cancer Lett. 2022; 530:45–58. DOI: 10.1016/j.canlet.2022.01.011. PMID: 35051533.
Article26. Zhang W, Su X, Li S, Liu Z, Wang Q, Zeng H. Low serum exosomal miR-484 expression predicts unfavorable prognosis in ovarian cancer. Cancer Biomark. 2020; 27(4):485–491. DOI: 10.3233/CBM-191123. PMID: 32065786.
Article27. Li H, Xia M, Zheng S, Lin Y, Yu T, Xie Y, et al. Cerebrospinal fluid exosomal microRNAs as biomarkers for diagnosing or monitoring the progression of non-small cell lung cancer with leptomeningeal metastases. Biotechnol Genet Eng Rev. 2023 Feb 28 [Epub]. DOI: 10.1080/02648725.2023.2183613.
Article28. Tomita Y, Suzuki K, Yamasaki S, Toriumi K, Miyashita M, Ando S, et al. Urinary exosomal microRNAs as predictive biomarkers for persistent psychotic-like experiences. Schizophrenia. 2023; 9(1):14. DOI: 10.1038/s41537-023-00340-5. PMID: 36906656. PMCID: PMC10008540.
Article29. Wang B, Xu Y, Wei Y, Lv L, Liu N, Lin R, et al. Human mesenchymal stem cell-derived exosomal microRNA-143 promotes apoptosis and suppresses cell growth in pancreatic cancer via target gene regulation. Front Genet. 2021; 12:581694. DOI: 10.3389/fgene.2021.581694. PMID: 33643376. PMCID: PMC7907650.
Article30. Ramanathan S, Douglas SR, Alexander GM, Shenoda BB, Barrett JE, Aradillas E, et al. Exosome microRNA signatures in patients with complex regional pain syndrome undergoing plasma exchange. J Transl Med. 2019; 17(1):81. DOI: 10.1186/s12967-019-1833-3. PMID: 30871575. PMCID: PMC6419338.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Next-generation sequencing analysis of exosomal microRNAs: Fusobacterium nucleatum regulates the expression profiling of exosomal microRNAs in human colorectal cancer cells
- A glance into the roles of microRNAs (exosomal and non-exosomal) in polycystic ovary syndrome
- Circulating Plasma and Exosomal microRNAs as Indicators of Drug-Induced Organ Injury in Rodent Models
- Role of MicroRNA in Visceral Pain
- Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma