Int J Stem Cells.
2024 Feb;17(1):15-29. 10.15283/ijsc23086.
Exploring the Molecular and Developmental Dynamics of Endothelial Cell Differentiation
- Affiliations
-
- 1Department of Bioengineering, University of Washington, Seattle, WA, USA
- 2Institute for Stem Cell and Regenerative Medicine, University of Washington, Seattle, WA, USA
- 3Department of Dermatology, School of Medicine, University of Washington, Seattle, WA, USA
- KMID: 2552014
- DOI: http://doi.org/10.15283/ijsc23086
Abstract
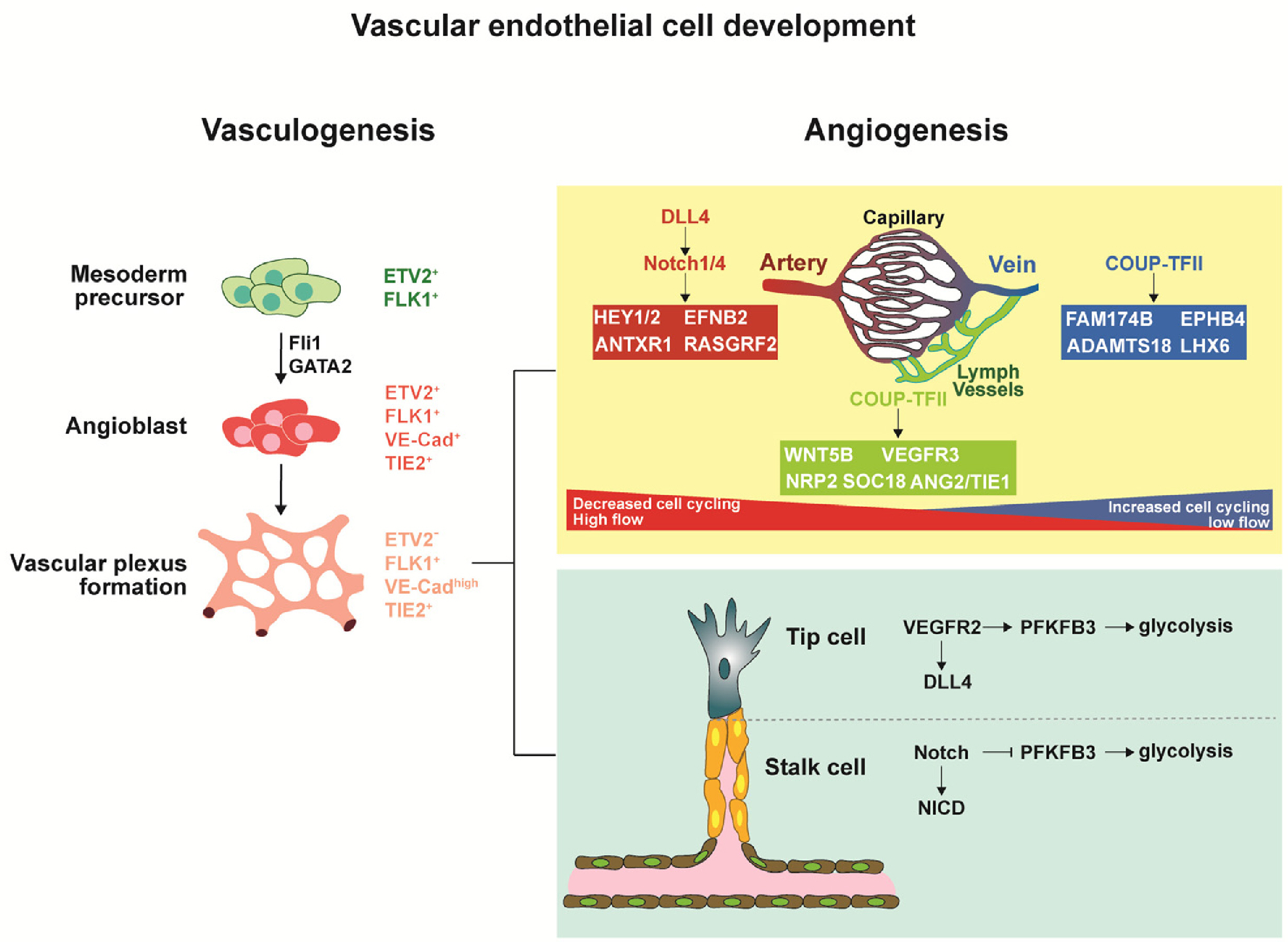
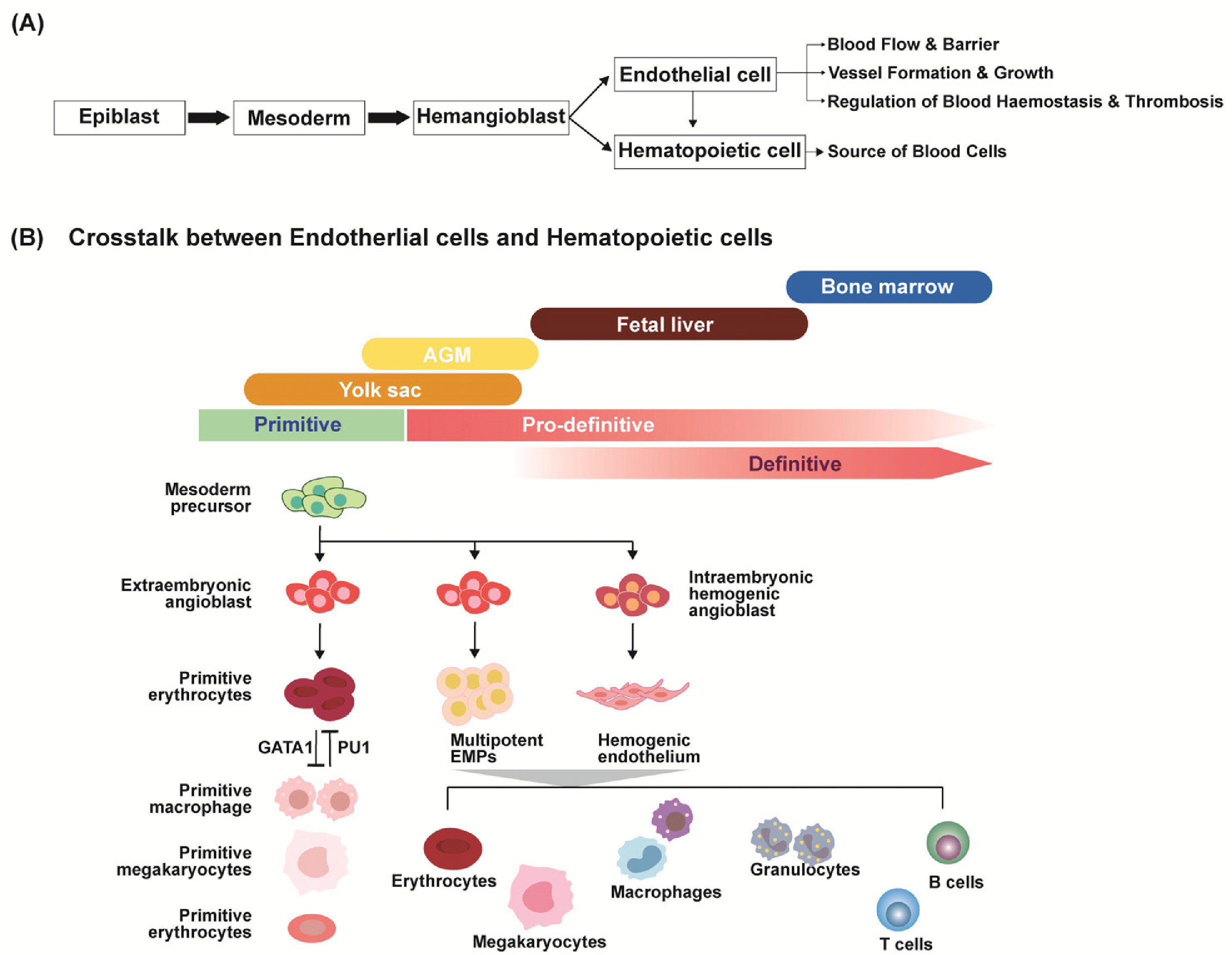
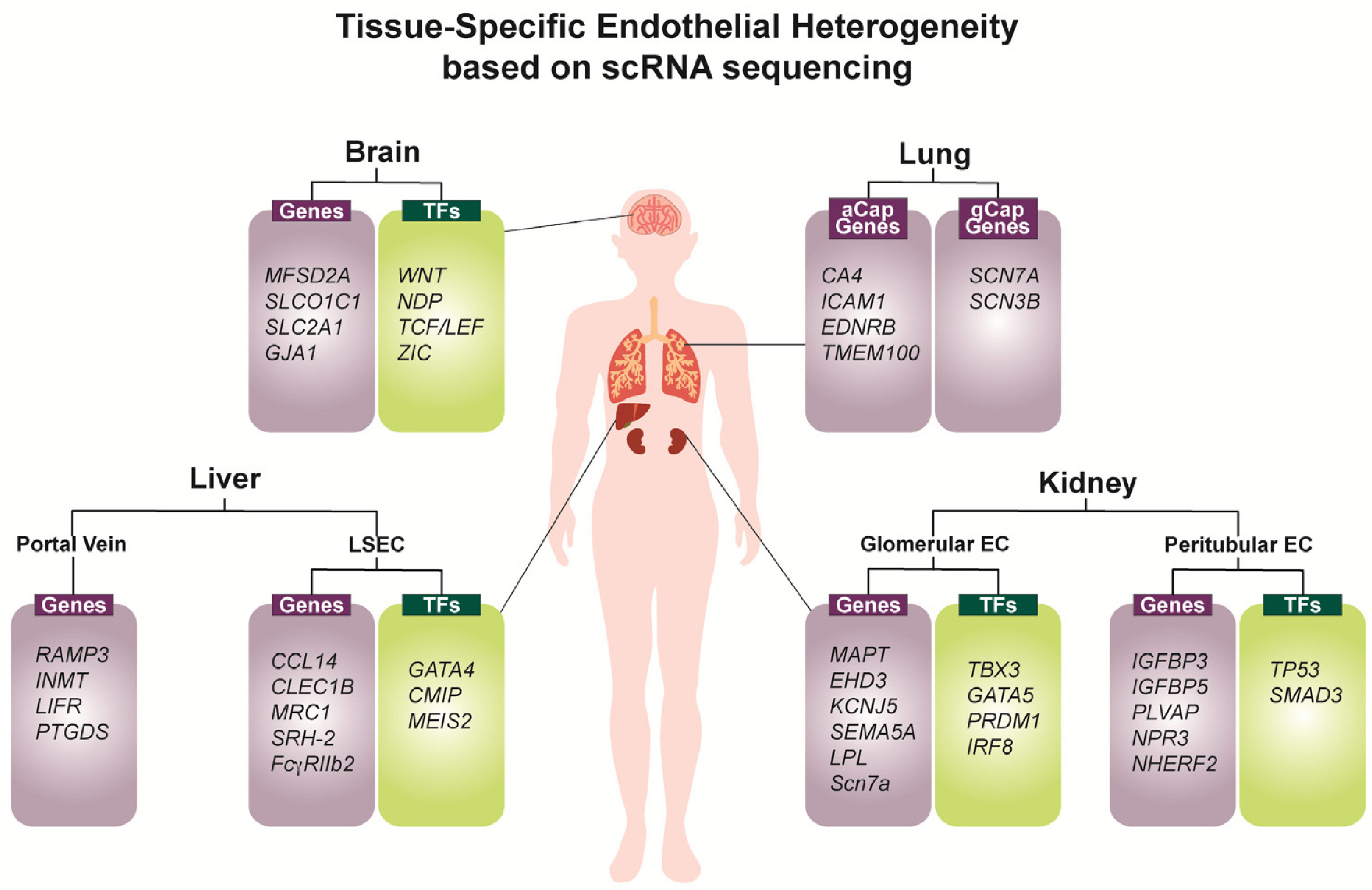
- The development and differentiation of endothelial cells (ECs) are fundamental processes with significant implications for both health and disease. ECs, which are found in all organs and blood vessels, play a crucial role in facilitating nutrient and waste exchange and maintaining proper vessel function. Understanding the intricate signaling pathways involved in EC development holds great promise for enhancing vascularization, tissue engineering, and vascular regeneration. Hematopoietic stem cells originating from hemogenic ECs, give rise to diverse immune cell populations, and the interaction between ECs and immune cells is vital for maintaining vascular integrity and regulating immune responses. Dysregulation of vascular development pathways can lead to various diseases, including cancer, where tumor-specific ECs promote tumor growth through angiogenesis. Recent advancements in single-cell genomics and in vivo genetic labeling have shed light on EC development, plasticity, and heterogeneity, uncovering tissue-specific gene expression and crucial signaling pathways. This review explores the potential of ECs in various applications, presenting novel opportunities for advancing vascular medicine and treatment strategies.
Figure
Reference
-
References
1. Qiu J, Hirschi KK. 2019; Endothelial cell development and its application to regenerative medicine. Circ Res. 125:489–501. DOI: 10.1161/CIRCRESAHA.119.311405. PMID: 31518171. PMCID: PMC8109152.
Article2. Carmeliet P, Ferreira V, Breier G, et al. 1996; Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 380:435–439. DOI: 10.1038/380435a0. PMID: 8602241.
Article3. Miquerol L, Langille BL, Nagy A. 2000; Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 127:3941–3946. DOI: 10.1242/dev.127.18.3941. PMID: 10952892.
Article4. Lee S, Chen TT, Barber CL, et al. 2007; Autocrine VEGF signaling is required for vascular homeostasis. Cell. 130:691–703. DOI: 10.1016/j.cell.2007.06.054. PMID: 17719546. PMCID: PMC3010851.
Article5. Gale NW, Dominguez MG, Noguera I, et al. 2004; Haploinsuffi-ciency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U S A. 101:15949–15954. DOI: 10.1073/pnas.0407290101. PMID: 15520367. PMCID: PMC524697.
Article6. Hellström M, Phng LK, Hofmann JJ, et al. 2007; Dll4 signalling through Notch1 regulates formation of tip cells during angio-genesis. Nature. 445:776–780. DOI: 10.1038/nature05571. PMID: 17259973.
Article7. Gerhardt H, Golding M, Fruttiger M, et al. 2003; VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 161:1163–1177. DOI: 10.1083/jcb.200302047. PMID: 12810700. PMCID: PMC2172999.
Article8. Cao Y, Zhang X, Wang L, et al. 2019; PFKFB3-mediated endothelial glycolysis promotes pulmonary hypertension. Proc Natl Acad Sci U S A. 116:13394–13403. DOI: 10.1073/pnas.1821401116. PMID: 31213542. PMCID: PMC6613097.
Article9. De Bock K, Georgiadou M, Schoors S, et al. 2013; Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 154:651–663. DOI: 10.1016/j.cell.2013.06.037. PMID: 23911327.
Article10. Stratman AN, Schwindt AE, Malotte KM, Davis GE. 2010; Endo-thelial-derived PDGF-BB and HB-EGF coordinately regulate pericyte recruitment during vasculogenic tube assembly and stabilization. Blood. 116:4720–4730. DOI: 10.1182/blood-2010-05-286872. PMID: 20739660. PMCID: PMC2996127.
Article11. Stenzel D, Nye E, Nisancioglu M, Adams RH, Yamaguchi Y, Gerhardt H. 2009; Peripheral mural cell recruitment requires cell-autonomous heparan sulfate. Blood. 114:915–924. DOI: 10.1182/blood-2008-10-186239. PMID: 19398718.
Article12. Hellström M, Gerhardt H, Kalén M, et al. 2001; Lack of pericytes leads to endothelial hyperplasia and abnormal vascular mor-phogenesis. J Cell Biol. 153:543–553. DOI: 10.1083/jcb.153.3.543. PMID: 11331305. PMCID: PMC2190573.
Article13. Eklund L, Kangas J, Saharinen P. 2017; Angiopoietin-Tie signalling in the cardiovascular and lymphatic systems. Clin Sci (Lond). 131:87–103. DOI: 10.1042/CS20160129. PMID: 27941161. PMCID: PMC5146956.
Article14. Fukuhara S, Sako K, Minami T, et al. 2008; Differential function of Tie2 at cell-cell contacts and cell-substratum contacts regulated by angiopoietin-1. Nat Cell Biol. 10:513–526. DOI: 10.1038/ncb1714. PMID: 18425120.
Article15. Brindle NP, Saharinen P, Alitalo K. 2006; Signaling and functions of angiopoietin-1 in vascular protection. Circ Res. 98:1014–1023. DOI: 10.1161/01.RES.0000218275.54089.12. PMID: 16645151. PMCID: PMC2270395.
Article16. Saharinen P, Eklund L, Alitalo K. 2017; Therapeutic targeting of the angiopoietin-TIE pathway. Nat Rev Drug Discov. 16:635–661. DOI: 10.1038/nrd.2016.278. PMID: 28529319.
Article17. Lobov IB, Brooks PC, Lang RA. 2002; Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proc Natl Acad Sci U S A. 99:11205–11210. DOI: 10.1073/pnas.172161899. PMID: 12163646. PMCID: PMC123234.
Article18. Liu ZJ, Shirakawa T, Li Y, et al. 2003; Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 23:14–25. DOI: 10.1128/MCB.23.1.14-25.2003. PMID: 12482957. PMCID: PMC140667.
Article19. Quillien A, Moore JC, Shin M, et al. 2014; Distinct Notch signaling outputs pattern the developing arterial system. Deve-lopment. 141:1544–1552. DOI: 10.1242/dev.099986. PMID: 24598161. PMCID: PMC4074308.
Article20. Coultas L, Nieuwenhuis E, Anderson GA, et al. 2010; Hedgehog regulates distinct vascular patterning events through VEGF-dependent and -independent mechanisms. Blood. 116:653–660. DOI: 10.1182/blood-2009-12-256644. PMID: 20339091.
Article21. Corada M, Nyqvist D, Orsenigo F, et al. 2010; The Wnt/beta-catenin pathway modulates vascular remodeling and specification by upregulating Dll4/Notch signaling. Dev Cell. 18:938–949. DOI: 10.1016/j.devcel.2010.05.006. PMID: 20627076. PMCID: PMC8127076.
Article22. Wythe JD, Dang LT, Devine WP, et al. 2013; ETS factors regulate Vegf-dependent arterial specification. Dev Cell. 26:45–58. DOI: 10.1016/j.devcel.2013.06.007. PMID: 23830865. PMCID: PMC3754838.
Article23. Stalmans I, Ng YS, Rohan R, et al. 2002; Arteriolar and venular patterning in retinas of mice selectively expressing VEGF isoforms. J Clin Invest. 109:327–336. DOI: 10.1172/JCI0214362. PMID: 11827992. PMCID: PMC150858.
Article24. Seo S, Kume T. 2006; Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol. 296:421–436. DOI: 10.1016/j.ydbio.2006.06.012. PMID: 16839542.
Article25. You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. 2005; Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 435:98–104. DOI: 10.1038/nature03511. PMID: 15875024.
Article26. le Noble F, Fleury V, Pries A, Corvol P, Eichmann A, Re-neman RS. 2005; Control of arterial branching morphogenesis in embryogenesis: go with the flow. Cardiovasc Res. 65:619–628. DOI: 10.1016/j.cardiores.2004.09.018. PMID: 15664388.
Article27. Peirce SM, Skalak TC. 2003; Microvascular remodeling: a complex continuum spanning angiogenesis to arteriogenesis. Microcir-culation. 10:99–111. DOI: 10.1080/713773592.
Article28. Fang JS, Coon BG, Gillis N, et al. 2017; Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun. 8:2149. Erratum in: Nat Commun 2018;9:720. DOI: 10.1038/s41467-018-03076-4. PMID: 29445140. PMCID: PMC5813030. PMID: 101eb7b7054e4d6f967907dee647da59.
Article29. Noseda M, Chang L, McLean G, et al. 2004; Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol Cell Biol. 24:8813–8822. DOI: 10.1128/MCB.24.20.8813-8822.2004. PMID: 15456857. PMCID: PMC517869.
Article30. Kao CY, Xu M, Wang L, et al. 2020; Elevated COUP-TFII expression in dopaminergic neurons accelerates the progre-ssion of Parkinson's disease through mitochondrial dysfun-ction. PLoS Genet. 16:e1008868. DOI: 10.1371/journal.pgen.1008868. PMID: 32579581. PMCID: PMC7340320. PMID: 48cc175e61f641c685c04c8ee0bf0d4f.
Article31. Xie X, Tang K, Yu CT, Tsai SY, Tsai MJ. 2013; Regulatory potential of COUP-TFs in development: stem/progenitor cells. Semin Cell Dev Biol. 24:687–693. DOI: 10.1016/j.semcdb.2013.08.005. PMID: 23978678. PMCID: PMC3849206.
Article32. Chavkin NW, Genet G, Poulet M, et al. 2022; Endothelial cell cycle state determines propensity for arterial-venous fate. Nat Commun. 13:5891. DOI: 10.1038/s41467-022-33324-7. PMID: 36202789. PMCID: PMC9537338. PMID: dd49956f70db4db48d1cedcdc042f6bb.
Article33. Dzierzak E, Bigas A. 2018; Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 22:639–651. DOI: 10.1016/j.stem.2018.04.015. PMID: 29727679.
Article34. Wu Y, Hirschi KK. 2021; Regulation of hemogenic endothelial cell development and function. Annu Rev Physiol. 83:17–37. DOI: 10.1146/annurev-physiol-021119-034352. PMID: 33035429. PMCID: PMC8634156.
Article35. Godin I, Cumano A. 2002; The hare and the tortoise: an embryonic haematopoietic race. Nat Rev Immunol. 2:593–604. DOI: 10.1038/nri857. PMID: 12154378.
Article36. Tober J, Koniski A, McGrath KE, et al. 2007; The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hema-topoiesis. Blood. 109:1433–1441. DOI: 10.1182/blood-2006-06-031898. PMID: 17062726. PMCID: PMC1794060.
Article37. Rekhtman N, Radparvar F, Evans T, Skoultchi AI. 1999; Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398–1411. DOI: 10.1101/gad.13.11.1398. PMID: 10364157. PMCID: PMC316770.
Article38. Frame JM, Fegan KH, Conway SJ, McGrath KE, Palis J. 2016; Definitive hematopoiesis in the yolk sac emerges from Wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells. 34:431–444. DOI: 10.1002/stem.2213. PMID: 26418893. PMCID: PMC4755868.
Article39. Ghosn E, Yoshimoto M, Nakauchi H, Weissman IL, Her-zenberg LA. 2019; Hematopoietic stem cell-independent hemato-poiesis and the origins of innate-like B lymphocytes. Deve-lopment. 146:dev170571. DOI: 10.1242/dev.170571. PMID: 31371526. PMCID: PMC6703711.
Article40. Ginhoux F, Greter M, Leboeuf M, et al. 2010; Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. DOI: 10.1126/science.1194637. PMID: 20966214. PMCID: PMC3719181.
Article41. Perdiguero EG, Klapproth K, Schulz C, et al. 2015; The origin of tissue-resident macrophages: when an erythro-myeloid progenitor is an erythro-myeloid progenitor. Immunity. 43:1023–1024. DOI: 10.1016/j.immuni.2015.11.022. PMID: 26682973.
Article42. Sheng J, Ruedl C, Karjalainen K. 2015; Most tissue-resident macrophages except microglia are derived from fetal hemato-poietic stem cells. Immunity. 43:382–393. DOI: 10.1016/j.immuni.2015.07.016. PMID: 26287683.
Article43. Perdiguero EG, Geissmann F. 2016; The development and maintenance of resident macrophages. Nat Immunol. 17:2–8. DOI: 10.1038/ni.3341. PMID: 26681456. PMCID: PMC4950995.
Article44. Lieu YK, Reddy EP. 2009; Conditional c-myb knockout in adult hematopoietic stem cells leads to loss of self-renewal due to impaired proliferation and accelerated differentiation. Proc Natl Acad Sci U S A. 106:21689–21694. DOI: 10.1073/pnas.0907623106. PMID: 19955420. PMCID: PMC2787467.
Article45. Motazedian A, Bruveris FF, Kumar SV, et al. 2020; Multipotent RAG1+ progenitors emerge directly from haemogenic endothelium in human pluripotent stem cell-derived haematopoietic organoids. Nat Cell Biol. 22:60–73. DOI: 10.1038/s41556-019-0445-8. PMID: 31907413.
Article46. Kobayashi M, Shelley WC, Seo W, et al. 2014; Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfβ for their develop-ment. Proc Natl Acad Sci U S A. 111:12151–12156. DOI: 10.1073/pnas.1407370111. PMID: 25092306. PMCID: PMC4143017.
Article47. Gritz E, Hirschi KK. 2016; Specification and function of hemogenic endothelium during embryogenesis. Cell Mol Life Sci. 73:1547–1567. DOI: 10.1007/s00018-016-2134-0. PMID: 26849156. PMCID: PMC4805691.
Article48. Müller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. 1994; Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1:291–301. DOI: 10.1016/1074-7613(94)90081-7. PMID: 7889417.
Article49. de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. 2000; Defini-tive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J. 19:2465–2474. DOI: 10.1093/emboj/19.11.2465. PMID: 10835345. PMCID: PMC212758.
Article50. Taoudi S, Medvinsky A. 2007; Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc Natl Acad Sci U S A. 104:9399–9403. DOI: 10.1073/pnas.0700984104. PMID: 17517650. PMCID: PMC1890506.
Article51. Fadlullah MZH, Neo WH, Lie-A-Ling M, et al. 2022; Murine AGM single-cell profiling identifies a continuum of hemogenic endothelium differentiation marked by ACE. Blood. 139:343–356. DOI: 10.1182/blood.2020007885. PMID: 34517413. PMCID: PMC9159109.
Article52. Gomes AM, Kurochkin I, Chang B, et al. 2018; Cooperative transcription factor induction mediates hemogenic reprogram-ming. Cell Rep. 25:2821–2835.e7. DOI: 10.1016/j.celrep.2018.11.032. PMID: 30517869. PMCID: PMC6571141.
Article53. Thambyrajah R, Patel R, Mazan M, et al. 2016; New insights into the regulation by RUNX1 and GFI1(s) proteins of the endothelial to hematopoietic transition generating primordial hematopoietic cells. Cell Cycle. 15:2108–2114. DOI: 10.1080/15384101.2016.1203491. PMID: 27399214. PMCID: PMC4993433.
Article54. Lancrin C, Mazan M, Stefanska M, et al. 2012; GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 120:314–322. DOI: 10.1182/blood-2011-10-386094. PMID: 22668850.
Article55. Bonkhofer F, Rispoli R, Pinheiro P, et al. 2019; Blood stem cell-forming haemogenic endothelium in zebrafish derives from arterial endothelium. Nat Commun. 10:3577. DOI: 10.1038/s41467-019-11423-2. PMID: 31395869. PMCID: PMC6687740. PMID: 7daa7a5352524efcafc9c49f85909b86.
Article56. Zhou Y, Zhang Y, Chen B, et al. 2019; Overexpression of GATA2 enhances development and maintenance of human embryonic stem cell-derived hematopoietic stem cell-like progeni-tors. Stem Cell Reports. 13:31–47. DOI: 10.1016/j.stemcr.2019.05.007. PMID: 31178416. PMCID: PMC6626852.
Article57. Abdelfattah A, Hughes-Davies A, Clayfield L, et al. 2021; Gata2 haploinsufficiency promotes proliferation and functional decline of hematopoietic stem cells with myeloid bias during aging. Blood Adv. 5:4285–4290. DOI: 10.1182/bloodadvances.2021004726. PMID: 34496012. PMCID: PMC8945642.
Article58. Coşkun S, Chao H, Vasavada H, et al. 2014; Development of the fetal bone marrow niche and regulation of HSC quiescence and homing ability by emerging osteolineage cells. Cell Rep. 9:581–590. DOI: 10.1016/j.celrep.2014.09.013. PMID: 25310984. PMCID: PMC4266564. PMID: 3c968bddcba44276b385be845dfbe5dc.
Article59. Braet F, Wisse E. 2002; Structural and functional aspects of liver sinusoidal endothelial cell fenestrae: a review. Comp Hepatol. 1:1. DOI: 10.1186/1476-5926-1-1. PMID: 12437787. PMCID: PMC131011.60. Kadry H, Noorani B, Cucullo L. 2020; A blood-brain barrier overview on structure, function, impairment, and biomar-kers of integrity. Fluids Barriers CNS. 17:69. DOI: 10.1186/s12987-020-00230-3. PMID: 33208141. PMCID: PMC7672931. PMID: 8955835654424d599d7df190b149dbde.
Article61. Jambusaria A, Hong Z, Zhang L, et al. 2020; Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. 9:e51413. DOI: 10.7554/eLife.51413. PMID: 31944177. PMCID: PMC7002042. PMID: 48432eae5df34dc8902eb012a4ba6d9b.
Article62. Schaum N, Lehallier B, Hahn O, et al. 2020; Ageing hallmarks exhibit organ-specific temporal signatures. Nature. 583:596–602. DOI: 10.1038/s41586-020-2499-y. PMID: 32669715. PMCID: PMC7757734.
Article63. Tabula Muris Consortium. Overall coordination. Logistical coordination. . 2018; Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 562:367–372. DOI: 10.1038/s41586-018-0590-4. PMID: 30283141. PMCID: PMC6642641.64. Paik DT, Tian L, Williams IM, et al. 2020; Single-cell RNA sequencing unveils unique transcriptomic signatures of organ-specific endothelial cells. Circulation. 142:1848–1862. DOI: 10.1161/CIRCULATIONAHA.119.041433. PMID: 32929989. PMCID: PMC7658053.
Article65. Marcu R, Choi YJ, Xue J, et al. 2018; Human organ-specific endothelial cell heterogeneity. iScience. 4:20–35. DOI: 10.1016/j.isci.2018.05.003. PMID: 30240741. PMCID: PMC6147238.
Article66. Kalucka J, de Rooij LPMH, Goveia J, et al. 2020; Single-cell transcriptome atlas of murine endothelial cells. Cell. 180:764–779.e20. DOI: 10.1016/j.cell.2020.01.015. PMID: 32059779.
Article67. Barry DM, McMillan EA, Kunar B, et al. 2019; Molecular determinants of nephron vascular specialization in the kidney. Nat Commun. 10:5705. DOI: 10.1038/s41467-019-12872-5. PMID: 31836710. PMCID: PMC6910926. PMID: 89ca827692df403aa71f8398b5ac7748.
Article68. Yang AC, Vest RT, Kern F, et al. 2022; A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature. 603:885–892. DOI: 10.1038/s41586-021-04369-3. PMID: 35165441. PMCID: PMC9635042.
Article69. Vanlandewijck M, He L, Mäe MA, et al. 2018; A molecular atlas of cell types and zonation in the brain vasculature. Nature. 554:475–480. Erratum in: Nature 2018;560:E3. DOI: 10.1038/nature25739. PMID: 29443965.
Article70. Inverso D, Shi J, Lee KH, et al. 2021; A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie-Wnt signaling axis in the liver. Dev Cell. 56:1677–1693.e10. DOI: 10.1016/j.devcel.2021.05.001. PMID: 34038707. PMCID: PMC8191494.
Article71. Sabbagh MF, Heng JS, Luo C, et al. 2018; Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. Elife. 7:e36187. DOI: 10.7554/eLife.36187. PMID: 30188322. PMCID: PMC6126923. PMID: 561fe078565b4184bf69573280bb6c71.
Article72. Genet N, Genet G, Chavkin NW, et al. 2023; Connexin 43-media-ted neurovascular interactions regulate neurogenesis in the adult brain subventricular zone. Cell Rep. 42:112371. DOI: 10.1016/j.celrep.2023.112371. PMID: 37043357. PMCID: PMC10564973.
Article73. Gillich A, Zhang F, Farmer CG, et al. 2020; Capillary cell-type specialization in the alveolus. Nature. 586:785–789. DOI: 10.1038/s41586-020-2822-7. PMID: 33057196. PMCID: PMC7721049.
Article74. Godoy RS, Cober ND, Cook DP, et al. 2023; Single-cell transcriptomic atlas of lung microvascular regeneration after targeted endothelial cell ablation. Elife. 12:e80900. DOI: 10.7554/eLife.80900. PMID: 37078698. PMCID: PMC10181823. PMID: 6be9dda738014335b2a97fbb476221b2.
Article75. Hua Y, Vella G, Rambow F, et al. 2022; Cancer immunotherapies transition endothelial cells into HEVs that generate TCF1+ T lymphocyte niches through a feed-forward loop. Cancer Cell. 40:1600–1618.e10. Erratum in: Cancer Cell 2023; 41:226. DOI: 10.1016/j.ccell.2022.11.002. PMID: 36423635. PMCID: PMC9899876.
Article76. De Smedt J, van Os EA, Talon I, et al. 2021; PU.1 drives specification of pluripotent stem cell-derived endothelial cells to LSEC-like cells. Cell Death Dis. 12:84. DOI: 10.1038/s41419-020-03356-2. PMID: 33446637. PMCID: PMC7809369. PMID: a72ccbfa4b2f428e822457e0c7616dda.
Article77. Halpern KB, Shenhav R, Massalha H, et al. 2018; Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 36:962–970. DOI: 10.1038/nbt.4231. PMID: 30222169. PMCID: PMC6546596.
Article78. Levy S, Sutton G, Ng PC, et al. 2007; The diploid genome sequence of an individual human. PLoS Biol. 5:e254. DOI: 10.1371/journal.pbio.0050254. PMID: 17803354. PMCID: PMC1964779. PMID: 1ebc1b9d3b354a3c848c72f1b19274b6.
Article79. Dumas SJ, Meta E, Borri M, et al. 2020; Single-cell RNA sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J Am Soc Nephrol. 31:118–138. DOI: 10.1681/ASN.2019080832. PMID: 31818909. PMCID: PMC6935008.80. Takemoto M, He L, Norlin J, et al. 2006; Large-scale identification of genes implicated in kidney glomerulus development and function. EMBO J. 25:1160–1174. DOI: 10.1038/sj.emboj.7601014. PMID: 16498405. PMCID: PMC1409724.
Article81. Wee H, Oh HM, Jo JH, Jun CD. 2009; ICAM-1/LFA-1 interaction contributes to the induction of endothelial cell-cell separation: implication for enhanced leukocyte diapedesis. Exp Mol Med. 41:341–348. DOI: 10.3858/emm.2009.41.5.038. PMID: 19307754. PMCID: PMC2701983.82. Gerszten RE, Luscinskas FW, Ding HT, et al. 1996; Adhesion of memory lymphocytes to vascular cell adhesion molecule-1-transduced human vascular endothelial cells under simulated physiological flow conditions in vitro. Circ Res. 79:1205–1215. DOI: 10.1161/01.RES.79.6.1205. PMID: 8943959.
Article83. Amersfoort J, Eelen G, Carmeliet P. 2022; Immunomodulation by endothelial cells - partnering up with the immune system? Nat Rev Immunol. 22:576–588. DOI: 10.1038/s41577-022-00694-4. PMID: 35288707. PMCID: PMC8920067.84. Wedgwood JF, Hatam L, Bonagura VR. 1988; Effect of interferon-gamma and tumor necrosis factor on the expression of class I and class II major histocompatibility molecules by cultured human umbilical vein endothelial cells. Cell Im-munol. 111:1–9. DOI: 10.1016/0008-8749(88)90046-9. PMID: 3123068.85. Limmer A, Ohl J, Kurts C, et al. 2000; Efficient presentation of exogenous antigen by liver endothelial cells to CD8+ T cells results in antigen-specific T-cell tolerance. Nat Med. 6:1348–1354. DOI: 10.1038/82161. PMID: 11100119.86. Zhao L, Li Z, Vong JSL, et al. 2020; Pharmacologically reversible zonation-dependent endothelial cell transcriptomic changes with neurodegenerative disease associations in the aged brain. Nat Commun. 11:4413. DOI: 10.1038/s41467-020-18249-3. PMID: 32887883. PMCID: PMC7474063. PMID: 0cd6b88e4d0b4038b2b9807cac7e8608.87. Shin YJ, Evitts KM, Jin S, et al. 2023; Amyloid beta peptides (Aβ) from Alzheimer's disease neuronal secretome induce endothelial activation in a human cerebral microvessel model. Neurobiol Dis. 181:106125. DOI: 10.1016/j.nbd.2023.106125. PMID: 37062307.88. Nascimento NR, Lessa LM, Kerntopf MR, et al. 2006; Inositols prevent and reverse endothelial dysfunction in diabetic rat and rabbit vasculature metabolically and by scavenging superoxide. Proc Natl Acad Sci U S A. 103:218–223. DOI: 10.1073/pnas.0509779103. PMID: 16373499. PMCID: PMC1325005.89. Kaludercic N, Di Lisa F. 2020; Mitochondrial ROS formation in the pathogenesis of diabetic cardiomyopathy. Front Cardiovasc Med. 7:12. DOI: 10.3389/fcvm.2020.00012. PMID: 32133373. PMCID: PMC7040199. PMID: 36f03c99410d4eaa8949a3fd4594561c.90. Mota RI, Morgan SE, Bahnson EM. 2020; Diabetic vasculopathy: macro and microvascular injury. Curr Pathobiol Rep. 8:1–14. DOI: 10.1007/s40139-020-00205-x. PMID: 32655983. PMCID: PMC7351096.91. Katakami N. 2018; Mechanism of development of atherosclerosis and cardiovascular disease in diabetes mellitus. J Athero-scler Thromb. 25:27–39. DOI: 10.5551/jat.RV17014. PMID: 28966336. PMCID: PMC5770221.92. Tacke F, Alvarez D, Kaplan TJ, et al. 2007; Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest. 117:185–194. DOI: 10.1172/JCI28549. PMID: 17200718. PMCID: PMC1716202.93. Li D, Mehta JL. 2000; Upregulation of endothelial receptor for oxidized LDL (LOX-1) by oxidized LDL and implications in apoptosis of human coronary artery endothelial cells: evidence from use of antisense LOX-1 mRNA and chemical inhibitors. Arterioscler Thromb Vasc Biol. 20:1116–1122. DOI: 10.1161/01.ATV.20.4.1116. PMID: 10764682.94. Davidson A, Aranow C. 2010; Lupus nephritis: lessons from murine models. Nat Rev Rheumatol. 6:13–20. DOI: 10.1038/nrrheum.2009.240. PMID: 19949431. PMCID: PMC4120882.
Article95. Renaudineau Y, Grunebaum E, Krause I, et al. 2001; Anti-endothelial cell antibodies (AECA) in systemic sclerosis--increased sensitivity using different endothelial cell substrates and association with other autoantibodies. Autoimmunity. 33:171–179. DOI: 10.3109/08916930109008045. PMID: 11683377.96. López-Isac E, Acosta-Herrera M, Kerick M, et al. 2019; GWAS for systemic sclerosis identifies multiple risk loci and highlights fibrotic and vasculopathy pathways. Nat Commun. 10:4955. DOI: 10.1038/s41467-019-12760-y. PMID: 31672989. PMCID: PMC6823490. PMID: 2c83b5b19dd94142a96da7824aa29e78.97. Benyamine A, Magalon J, Sabatier F, et al. 2018; Natural killer cells exhibit a peculiar phenotypic profile in systemic sclerosis and are potent inducers of endothelial microparticles release. Front Immunol. 9:1665. DOI: 10.3389/fimmu.2018.01665. PMID: 30072999. PMCID: PMC6058015. PMID: d92ef8da39dc45168134e54589c900b6.98. Folkman J. 1971; Tumor angiogenesis: therapeutic implications. N Engl J Med. 285:1182–1186. DOI: 10.1056/NEJM197111182852108. PMID: 4938153.99. Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. 1998; Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 92:735–745. DOI: 10.1016/S0092-8674(00)81402-6. PMID: 9529250.
Article100. Brady J, Neal J, Sadakar N, Gasque P. 2004; Human endosialin (tumor endothelial marker 1) is abundantly expressed in highly malignant and invasive brain tumors. J Neuropathol Exp Neurol. 63:1274–1283. DOI: 10.1093/jnen/63.12.1274. PMID: 15624764.101. Harjunpää H, Llort Asens M, Guenther C, Fagerholm SC. 2019; Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Front Immunol. 10:1078. DOI: 10.3389/fimmu.2019.01078. PMID: 31231358. PMCID: PMC6558418. PMID: 8d1781bf958940c398daf0b2b5e011a3.102. St Croix B, Rago C, Velculescu V, et al. 2000; Genes expressed in human tumor endothelium. Science. 289:1197–1202. DOI: 10.1126/science.289.5482.1197. PMID: 10947988.103. Bergers G, Benjamin LE. 2003; Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 3:401–410. DOI: 10.1038/nrc1093. PMID: 12778130.104. Guo P, Hu B, Gu W, et al. 2003; Platelet-derived growth factor-B enhances glioma angiogenesis by stimulating vascular endothelial growth factor expression in tumor endothelia and by promoting pericyte recruitment. Am J Pathol. 162:1083–1093. DOI: 10.1016/S0002-9440(10)63905-3. PMID: 12651601. PMCID: PMC1851242.
Article105. Cao Y, Cao R, Hedlund EM. 2008; R Regulation of tumor angiogenesis and metastasis by FGF and PDGF signaling path-ways. J Mol Med (Berl). 86:785–789. DOI: 10.1007/s00109-008-0337-z. PMID: 18392794.
Article106. Song M, Finley SD. 2020; ERK and Akt exhibit distinct signaling responses following stimulation by pro-angiogenic factors. Cell Commun Signal. 18:114. DOI: 10.1186/s12964-020-00595-w. PMID: 32680529. PMCID: PMC7368799. PMID: e5ba9e6d5a7c42698f96aeb699b9050d.107. Quintero-Fabián S, Arreola R, Becerril-Villanueva E, et al. 2019; Role of matrix metalloproteinases in angiogenesis and cancer. Front Oncol. 9:1370. DOI: 10.3389/fonc.2019.01370. PMID: 31921634. PMCID: PMC6915110. PMID: af6f90a4499b46a9bc6286338cefdcb4.108. Pouysségur J, Dayan F, Mazure NM. 2006; Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 441:437–443. DOI: 10.1038/nature04871. PMID: 16724055.
Article109. Waldman AD, Fritz JM, Lenardo MJ. 2020; A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 20:651–668. DOI: 10.1038/s41577-020-0306-5. PMID: 32433532. PMCID: PMC7238960.110. Zhang Y, Zhang Z. 2020; The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implica-tions. Cell Mol Immunol. 17:807–821. DOI: 10.1038/s41423-020-0488-6. PMID: 32612154. PMCID: PMC7395159.
Article111. Johnson DB, Nebhan CA, Moslehi JJ, Balko JM. 2022; Immune-checkpoint inhibitors: long-term implications of toxicity. Nat Rev Clin Oncol. 19:254–267. DOI: 10.1038/s41571-022-00600-w. PMID: 35082367. PMCID: PMC8790946.112. Johnson PC, Gainor JF, Sullivan RJ, Longo DL, Chabner B. 2023; Immune checkpoint inhibitors - the need for innovation. N Engl J Med. 388:1529–1532. DOI: 10.1056/NEJMsb2300232. PMID: 37075146.113. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. 2018; Immune checkpoint inhibitors: recent progress and potential bio-markers. Exp Mol Med. 50:1–11. DOI: 10.1038/s12276-018-0191-1. PMID: 30546008. PMCID: PMC6292890. PMID: 02cbef34102b4e70b2092a2f86e732c4.
Article114. Schaaf MB, Garg AD, Agostinis P. 2018; Defining the role of the tumor vasculature in antitumor immunity and immuno-therapy. Cell Death Dis. 9:115. DOI: 10.1038/s41419-017-0061-0. PMID: 29371595. PMCID: PMC5833710.115. Duru G, van Egmond M, Heemskerk N. 2020; A window of oppo-rtunity: targeting cancer endothelium to enhance immuno-therapy. Front Immunol. 11:584723. DOI: 10.3389/fimmu.2020.584723. PMID: 33262763. PMCID: PMC7686513. PMID: 013f6d9a138d45a5893baa1ee0c46533.
Article116. Griffioen AW, Damen CA, Blijham GH, Groenewegen G. 1996; Tumor angiogenesis is accompanied by a decreased infla-mmatory response of tumor-associated endothelium. Blood. 88:667–673. DOI: 10.1182/blood.V88.2.667.bloodjournal882667. PMID: 8695814.
Article117. Molon B, Ugel S, Del Pozzo F, et al. 2011; Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J Exp Med. 208:1949–1962. DOI: 10.1084/jem.20101956. PMID: 21930770. PMCID: PMC3182051.
Article118. Motz GT, Santoro SP, Wang LP, et al. 2014; Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat Med. 20:607–615. DOI: 10.1038/nm.3541. PMID: 24793239. PMCID: PMC4060245.
Article119. Rodig N, Ryan T, Allen JA, et al. 2003; Endothelial expression of PD-L1 and PD-L2 down-regulates CD8+ T cell activation and cytolysis. Eur J Immunol. 33:3117–3126. DOI: 10.1002/eji.200324270. PMID: 14579280.120. Lanitis E, Irving M, Coukos G. 2015; Targeting the tumor vasculature to enhance T cell activity. Curr Opin Immunol. 33:55–63. DOI: 10.1016/j.coi.2015.01.011. PMID: 25665467. PMCID: PMC4896929.121. Allen E, Jabouille A, Rivera LB, et al. 2017; Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci Transl Med. 9:eaak9679. DOI: 10.1126/scitranslmed.aak9679. PMID: 28404866. PMCID: PMC5554432.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Endothelial cell autophagy in the context of disease development
- Non-invasive molecular imaging of immune cell dynamics for vaccine research
- Cardiac side population cells exhibit endothelial differentiation potential
- Assessment of Developmental Toxicants using Human Embryonic Stem Cells
- Advances in Understanding the Molecular Biology of Brain Tumors