Ann Lab Med.
2023 Jul;43(4):392-394. 10.3343/alm.2023.43.4.392.
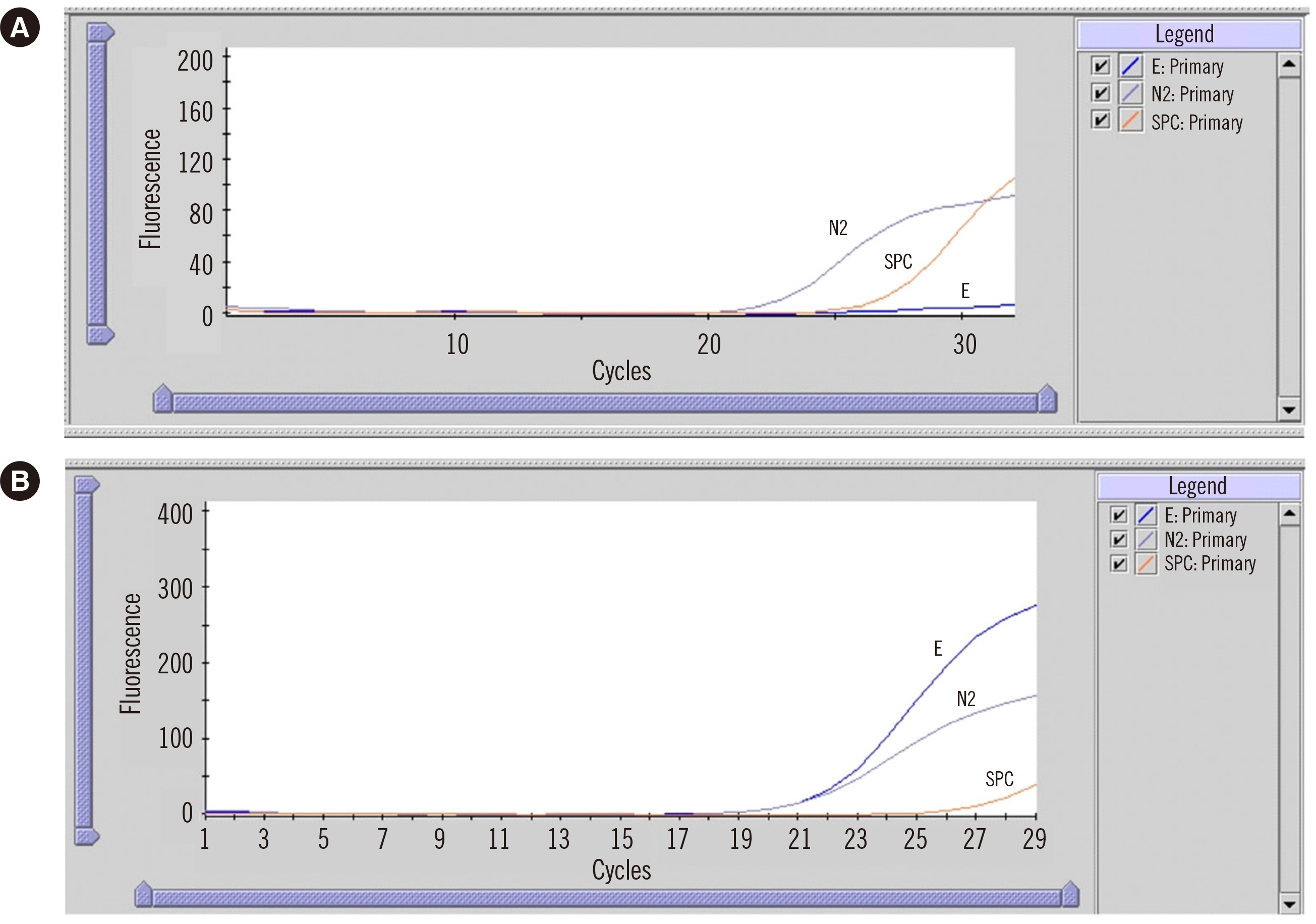
Deciphering a Case of SARS-CoV-2 E Gene Dropout With the Xpert Xpress Assay
- Affiliations
-
- 1Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Sanggye Paik Hospital, School of Medicine, Inje University, Seoul, Korea
- KMID: 2551959
- DOI: http://doi.org/10.3343/alm.2023.43.4.392
Figure
Reference
-
1. Kim YJ, Sung H, Ki CS, Hur M. 2020; COVID-19 testing in South Korea: current status and the need for faster diagnostics. Ann Lab Med. 40:349–50. DOI: 10.3343/alm.2020.40.5.349. PMID: 32237287. PMCID: PMC7169622.
Article2. Tahan S, Parikh BA, Droit L, Wallace MA, Burnham CD, Wang D. 2021; SARS-CoV-2 E gene variant alters analytical sensitivity characteristics of viral detection using a commercial reverse transcription-PCR assay. J Clin Microbiol. 59:e0007521. DOI: 10.1128/JCM.00075-21. PMID: 33903167. PMCID: PMC8218754.
Article3. Vanaerschot M, Mann SA, Webber JT, Kamm J, Bell SM, Bell J, et al. 2020; Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by reverse transcription-PCR. J Clin Microbiol. 59:e02369–20. DOI: 10.1128/JCM.02369-20. PMID: 33067272. PMCID: PMC7771441.
Article4. Rhoads DD, Plunkett D, Nakitandwe J, Dempsey A, Tu ZJ, Procop GW, et al. 2021; Endemic SARS-CoV-2 polymorphisms can cause a higher diagnostic target failure rate than estimated by aggregate global sequencing data. J Clin Microbiol. 59:e0091321. DOI: 10.1128/JCM.00913-21. PMID: 33972454. PMCID: PMC8288267.
Article5. Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. 2020; SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 25:2001650. DOI: 10.2807/1560-7917.ES.2020.25.39.2001650. PMID: 33006300. PMCID: PMC7531073.
Article6. Hong KH, In JW, Lee J, Kim SY, Lee KA, Kim S, et al. 2022; Prevalence of a single-nucleotide variant of SARS-CoV-2 in Korea and its impact on the diagnostic sensitivity of the Xpert Xpress SARS-CoV-2 assay. Ann Lab Med. 42:96–9. DOI: 10.3343/alm.2022.42.1.96. PMID: 34374354. PMCID: PMC8368233.
Article7. Schizas N, Michailidis T, Samiotis I, Patris V, Papakonstantinou K, Argiriou M, et al. 2020; Delayed diagnosis and treatment of a critically ill patient with infective endocarditis due to a false-positive molecular diagnostic test for SARS-CoV-2. Am J Case Rep. 21:e925931-1-3. DOI: 10.12659/AJCR.925931. PMID: 32980852. PMCID: PMC7526942.
Article8. Kanji JN, Zelyas N, MacDonald C, Pabbaraju K, Khan MN, Prasad A, et al. 2021; False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 18:13. DOI: 10.1186/s12985-021-01489-0. PMID: 33422083. PMCID: PMC7794619.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Retrospective Validation of Xpert Xpress SARSCoV-2 and Rapid COVID-19 PCR Tests
- Experiences of Preoperative COVID-19 Screening in Pediatric Patients in a Secondary Care Center
- Comparison of the Clinical Performance of the Point-of-care STANDARD M10 SARS-CoV-2 and Xpert Xpress SARS-CoV-2 Assays
- Performance of STANDARD™ M10 SARS-CoV-2 Assay for the Diagnosis of COVID-19 from a Nasopharyngeal Swab
- Clinical Usefulness of SARS-CoV-2 Antibody Test