Ann Lab Med.
2023 Jan;43(1):64-72. 10.3343/alm.2023.43.1.64.
Comparison of Homologous Recombination Repair Gene Next-Generation Sequencing Analysis in Patients With Metastatic Castration-Resistant Prostate Cancer Between Local and Central Laboratories in Korea
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea
- 2Center for Precision Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 3Department of Pathology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 4Department of Laboratory Medicine, Gangnam Severance Hospital, Seoul, Korea
- KMID: 2551587
- DOI: http://doi.org/10.3343/alm.2023.43.1.64
Abstract
- Background
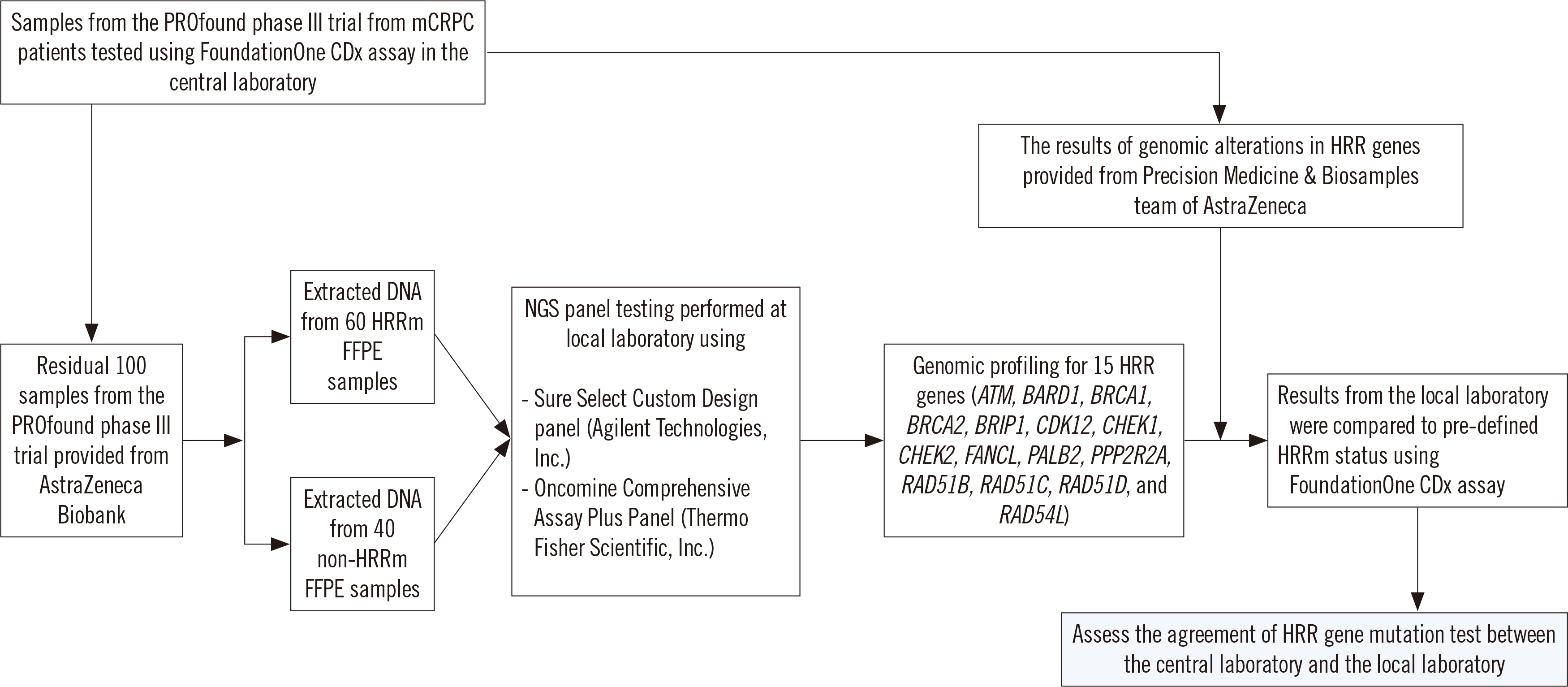
Following success of the phase III PROfound trial, the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib was approved by the US Food and Drug Administration in May 2020 for adult patients with deleterious homologous recombination repair (HRR) gene-mutated metastatic castration-resistant prostate cancer (mCRPC). As locally adopted multigene panel next-generation sequencing (NGS) assays for selecting PARP inhibitor candidates have not been thoroughly evaluated, we compared the analytical performance of the FoundationOne CDx (Foundation Medicine, Inc., Cambridge, MA, USA) (central laboratory) and other NGS assays (local laboratory) with samples from the PROfound trial in Korea.
Methods
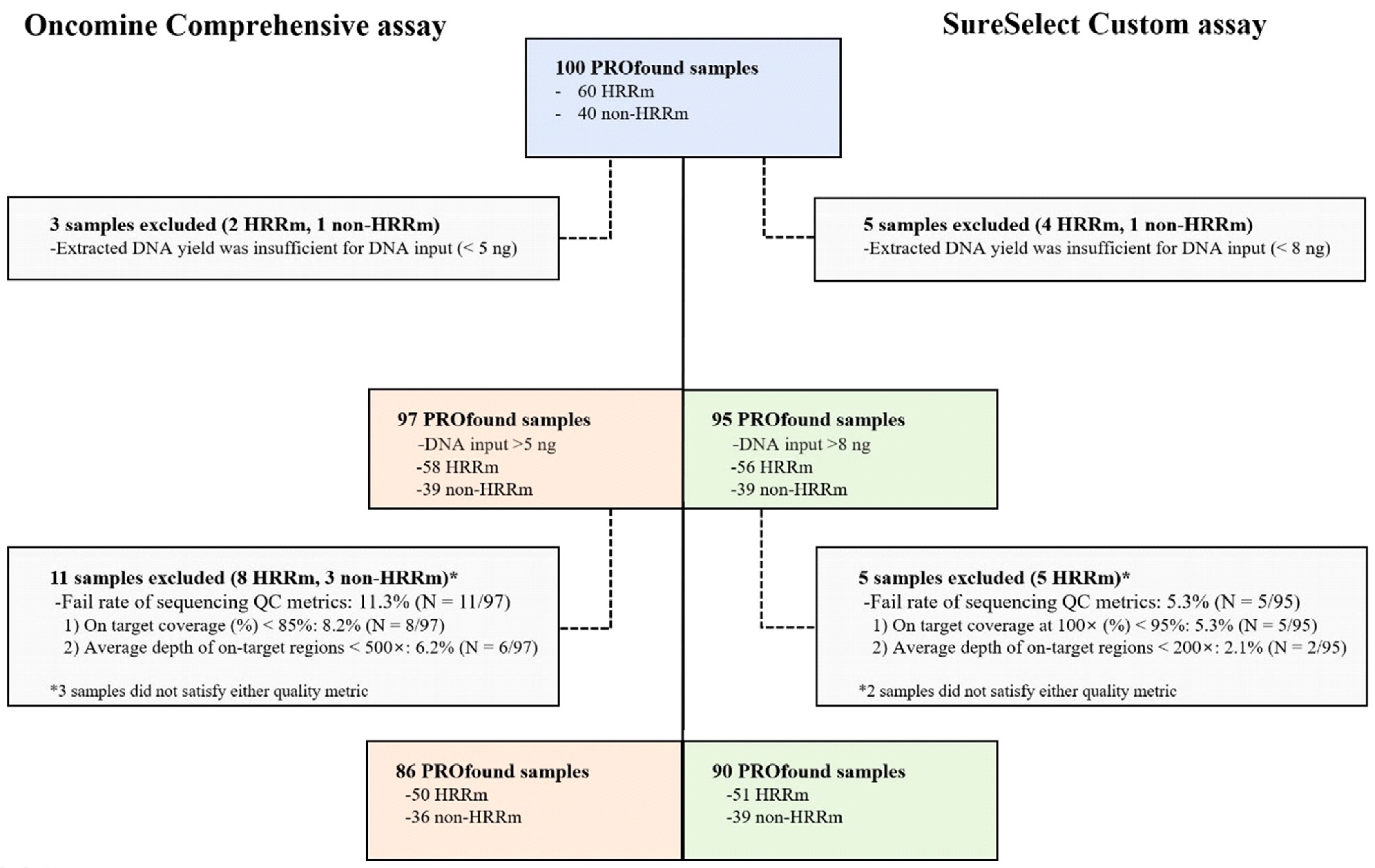
One hundred PROfound samples (60 HRR mutation [HRRm] cases and 40 non-HRRm cases) were analyzed. The results of HRR gene mutation analysis were compared between the FoundationOne CDx and two other NGS assays [SureSelect Custom Design assay (Agilent Technologies, Inc., Santa Clara, CA, USA) and Oncomine Comprehensive assay (Thermo Fisher Scientific, Inc., Waltham, MA, USA)].
Results
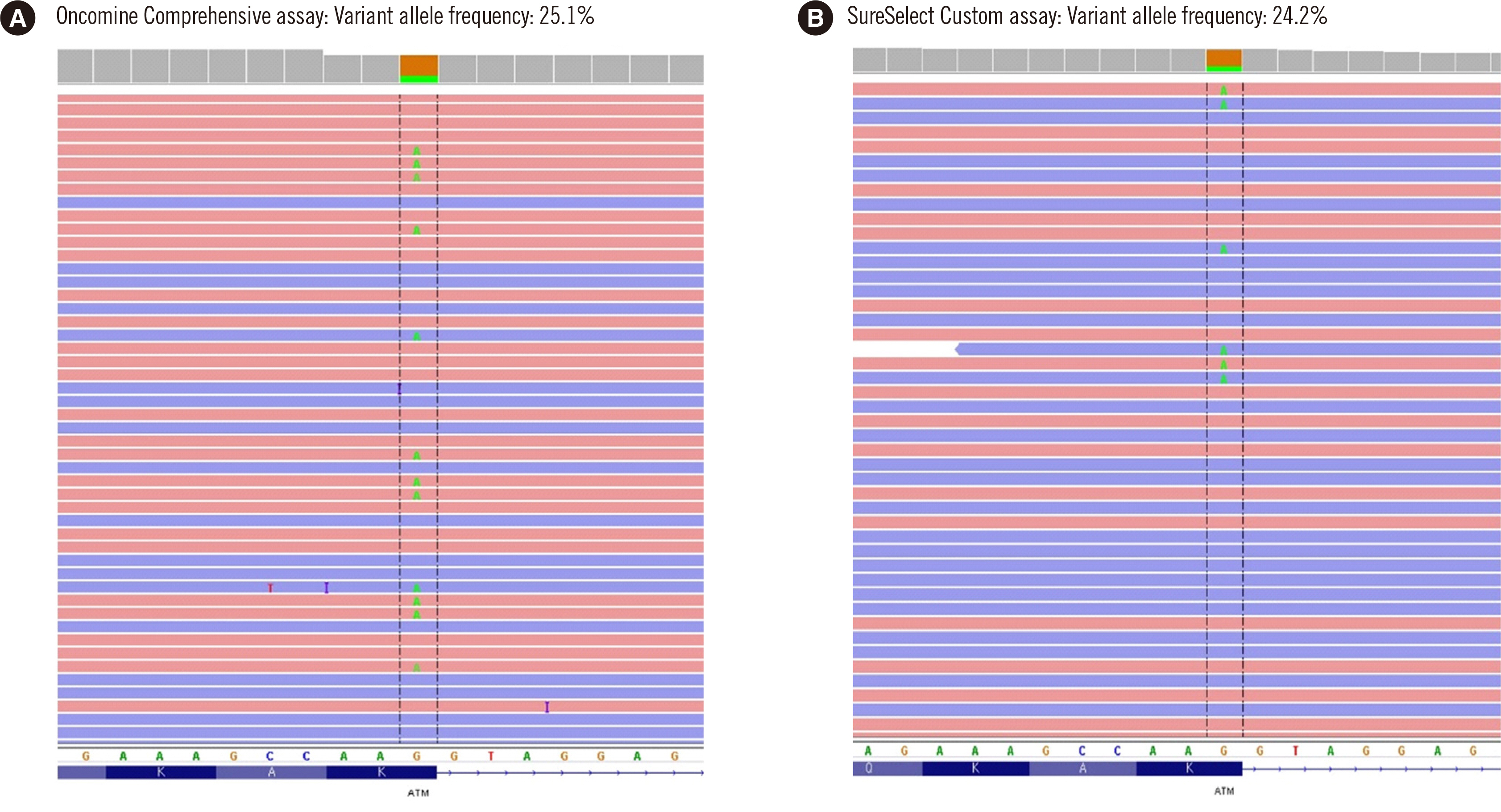
The positive percent agreement for single nucleotide variants (SNVs) and insertion/deletions (indels) between the central laboratory and local laboratory was 98.7%–100.0%. The negative percent agreement and overall percent agreement (OPA) for SNVs and indels between central and local laboratories were both 100%. Compared with that of the FoundationOne CDx assay, the OPA for copy number variations of the Oncomine Comprehensive and SureSelect Custom assays reached 99.8%–100%. Most mCRPC patients harboring a deleterious genetic variant were successfully identified with both local laboratory assays.
Conclusions
The NGS approach at a local laboratory showed comparable analytical performance for identifying HRRm status to the FoundationOne CDx assay used at the central laboratory.
Keyword
Figure
Reference
-
1. Adashek JJ, Jain RK, Zhang J. 2019; Clinical development of PARP inhibitors in treating metastatic castration-resistant prostate cancer. Cells. 8:860. DOI: 10.3390/cells8080860. PMID: 31404966. PMCID: PMC6721701.
Article2. Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. 2012; Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72:5588–99. DOI: 10.1158/0008-5472.CAN-12-2753. PMID: 23118055. PMCID: PMC3528345.
Article3. Ryan CJ, Abida W, Bryce AH, Balar AV, Dumbadze I, Given RW, et al. 2018; TRITON3: an international, randomized, open-label, phase III study of the PARP inhibitor rucaparib vs. physician's choice of therapy for patients with metastatic castration-resistant prostate cancer (mCRPC) associated with homologous recombination deficiency (HRD). J Clin Oncol. 36:TPS389. DOI: 10.1200/JCO.2018.36.6_suppl.TPS389.
Article4. Sigorski D, Iżycka-Świeszewska E, Bodnar L. 2020; Poly(ADP-ribose) polymerase inhibitors in prostate cancer: molecular mechanisms, and preclinical and clinical data. Target Oncol. 15:709–22. DOI: 10.1007/s11523-020-00756-4. PMID: 33044685. PMCID: PMC7701127.
Article5. Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. 2016; Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 375:443–53. DOI: 10.1056/NEJMoa1603144. PMID: 27433846. PMCID: PMC4986616.6. Cancer Genome Atlas Research Network. 2015; The molecular taxonomy of primary prostate cancer. Cell. 163:1011–25. DOI: 10.1016/j.cell.2015.10.025. PMID: 26544944. PMCID: PMC4695400.7. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. 2020; Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 382:2091–102. DOI: 10.1056/NEJMoa1911440. PMID: 32343890.
Article8. Garje R, Vaddepally RK, Zakharia Y. 2020; PARP inhibitors in prostate and urothelial cancers. Front Oncol. 10:114. DOI: 10.3389/fonc.2020.00114. PMID: 32117762. PMCID: PMC7020773.
Article9. Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, et al. 2012; A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 28:2747–54. DOI: 10.1093/bioinformatics/bts526. PMID: 22942019. PMCID: PMC3476336.
Article10. Tatsi C, Pankratz N, Lane J, Faucz FR, Hernández-Ramírez LC, Keil M, et al. 2019; Large genomic aberrations in corticotropinomas are associated with greater aggressiveness. J Clin Endocrinol Metab. 104:1792–801. DOI: 10.1210/jc.2018-02164. PMID: 30597087. PMCID: PMC6452317.
Article11. CLSI. 2008. User protocol for evaluation of qualitative test performance. 2nd ed. CLSI EP12-A2. Clinical and Laboratory Standards Institute;Wayne, PA: DOI: 10.1056/nejmc2023199.12. Khodakov D, Wang C, Zhang DY. 2016; Diagnostics based on nucleic acid sequence variant profiling: PCR, hybridization, and NGS approaches. Adv Drug Deliv Rev. 105:3–19. DOI: 10.1016/j.addr.2016.04.005. PMID: 27089811.
Article13. Seong MW, Kim M, Shin HS, Cho SI, Park SS. 2020; Three-year experience of an external proficiency testing survey for next-generation sequencing-based testing for germline mutation. Lab Med Qual Assur. 42:48–53. DOI: 10.15263/jlmqa.2020.42.1.48.
Article14. Zakrzewski F, Gieldon L, Rump A, Seifert M, Grützmann K, Krüger A, et al. 2019; Targeted capture-based NGS is superior to multiplex PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE tumor samples. BMC Cancer. 19:396. DOI: 10.1186/s12885-019-5584-6. PMID: 31029168. PMCID: PMC6487025.
Article15. Castro E, Mateo J, Olmos D, de Bono JS. 2016; Targeting DNA repair: the role of PARP inhibition in the treatment of castration-resistant prostate cancer. Cancer J. 22:353–6. DOI: 10.1097/PPO.0000000000000219. PMID: 27749330.16. Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, et al. 2018; Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018:PO.17.00286. DOI: 10.1200/PO.17.00286. PMID: 30234181. PMCID: PMC6139373.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Systemic Therapies for Metastatic Castration-Resistant Prostate Cancer: An Updated Review
- Next-Generation Sequencing in Prostate Cancer
- Chemotherapy With Androgen Deprivation for Hormone-Naïve Prostate Cancer
- Guidelines for Evaluating Treatment Response Based on Bone Scan for Metastatic Castration-Resistant Prostate Cancer: Prostate Cancer Clinical Trial Working Group 3 Recommendations
- Nonmetastatic Castration-Resistant Prostate Cancer