Cancer Res Treat.
2024 Jan;56(1):81-91. 10.4143/crt.2023.408.
Clinical Validation of the Unparalleled Sensitivity of the Novel Allele-Discriminating Priming System Technology–Based EGFR Mutation Assay in Patients with Operable Non–Small Cell Lung Cancer
- Affiliations
-
- 1GENECAST, Seoul, Korea
- 2Division of Data Science, Data Science Convergence Research Center, Hallym University, Chuncheon, Korea
- 3Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 4Department of Thoracic and Cardiovascular Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2550325
- DOI: http://doi.org/10.4143/crt.2023.408
Abstract
- Purpose
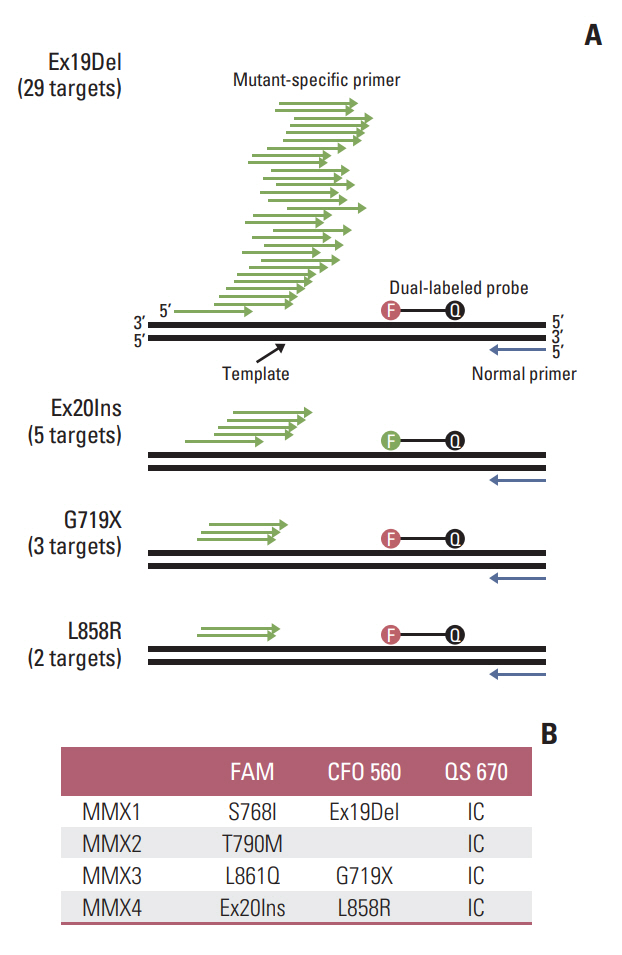
Recently, we developed allele-discriminating priming system (ADPS) technology. This method increases the sensitivity of conventional quantitative polymerase chain reaction up to 100 folds, with limit of detection, 0.01%, with reinforced specificity. This prospective study aimed to develop and validate the accuracy of ADPS epidermal growth factor receptor (EGFR) Mutation Test Kit using clinical specimens.
Materials and Methods
In total 189 formalin-fixed paraffin-embedded tumor tissues resected from patients with non–small cell lung cancer were used to perform a comparative evaluation of the ADPS EGFR Mutation Test Kit versus the cobas EGFR Mutation Test v2, which is the current gold standard. When the two methods had inconsistent results, next-generation sequencing–based CancerSCAN was utilized as a referee.
Results
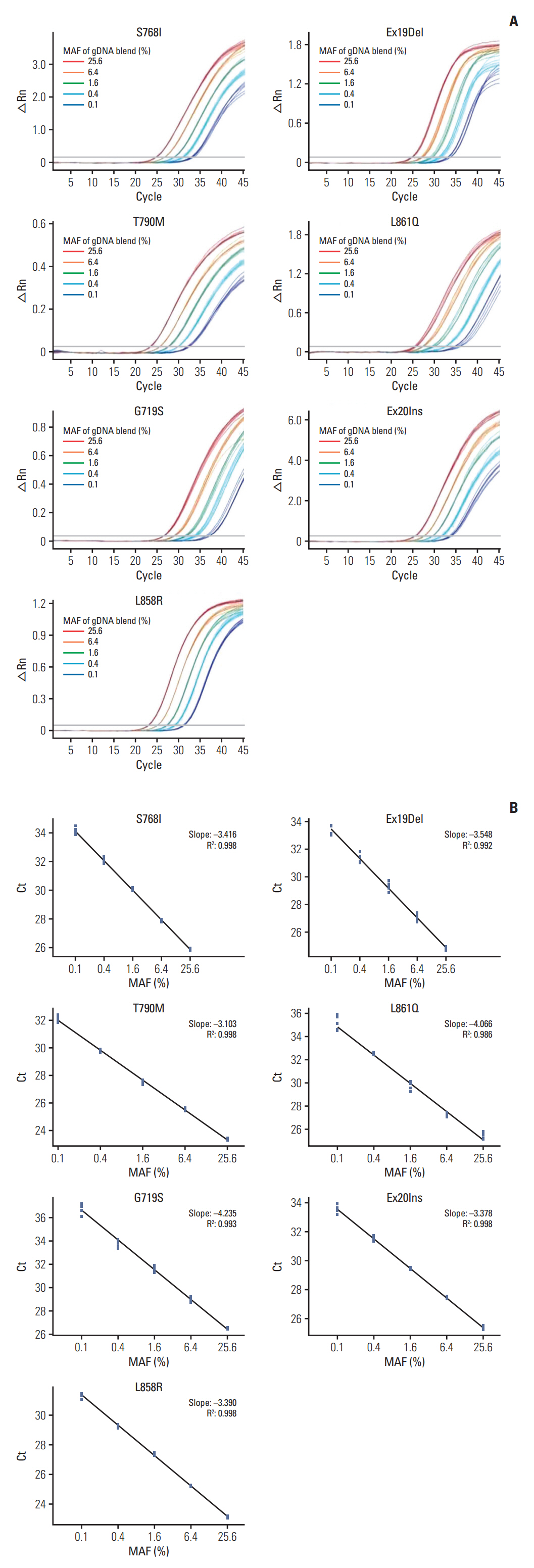
The overall agreement of the two methods was 97.4% (93.9%-99.1%); the positive percent agreement, 95.0% (88.7%-98.4%); and the negative percent agreement, 100.0% (95.9%-100.0%). EGFR mutations were detected at a frequency of 50.3% using the ADPS EGFR Mutation Test Kit and 52.9% using the cobas EGFR Mutation Test v2. There were 10 discrepant mutation calls between the two methods. CancerSCAN reproduced eight ADPS results. In two cases, mutant allele fraction was ultra-low at 0.02% and 0.06%, which are significantly below the limit of detection of the cobas assay and CancerSCAN. Based on the EGFR genotyping by ADPS, the treatment options could be switched in five patients.
Conclusion
The highly sensitive and specific ADPS EGFR Mutation Test Kit would be useful in detecting the patients who have lung cancer with EGFR mutation, and can benefit from the EGFR targeted therapy.
Keyword
Figure
Reference
-
References
1. Mok TS. Personalized medicine in lung cancer: what we need to know. Nat Rev Clin Oncol. 2011; 8:661–8.2. Hensing T, Chawla A, Batra R, Salgia R. A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol. 2014; 799:85–117.3. Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018; 19:269–85.4. Li X, Zhou C. Comparison of cross-platform technologies for EGFR T790M testing in patients with non-small cell lung cancer. Oncotarget. 2017; 8:100801–18.5. Nakamura H, Koizumi H, Sakai H, Kimura H, Miyazawa T, Marushima H, et al. Accuracy of the cobas EGFR mutation assay in non-small-cell lung cancer compared with three laboratory-developed tests. Clin Lung Cancer. 2018; 19:170–4.6. Syed YY. therascreen(R) EGFR RGQ PCR Kit: A companion diagnostic for afatinib and gefitinib in non-small cell lung cancer. Mol Diagn Ther. 2016; 20:191–8.7. Kim SS, Choi HJ, Kim JJ, Kim MS, Lee IS, Byun B, et al. Droplet digital PCR-based EGFR mutation detection with an internal quality control index to determine the quality of DNA. Sci Rep. 2018; 8:543.8. Shin HT, Choi YL, Yun JW, Kim NK, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017; 8:1377.9. Lee C, Bae JS, Ryu GH, Kim NKD, Park D, Chung J, et al. A method to evaluate the quality of clinical gene-panel sequencing data for single-nucleotide variant detection. J Mol Diagn. 2017; 19:651–8.10. Huang JK, Fan L, Wang TY, Wu PS. A new primer construction technique that effectively increases amplification of rare mutant templates in samples. BMC Biotechnol. 2019; 19:62.11. Kramer FR, Vargas DY. SuperSelective primer pairs for sensitive detection of rare somatic mutations. Sci Rep. 2021; 11:22384.12. Lim Y, Park IH, Lee HH, Baek K, Lee BC, Cho G. Modified Taq DNA polymerase for allele-specific ultra-sensitive detection of genetic variants. J Mol Diagn. 2022; 24:1128–42.13. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012; 13:239–46.14. Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013; 31:3327–34.15. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018; 378:113–25.16. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2020; 382:41–50.17. Kancha RK, von Bubnoff N, Peschel C, Duyster J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009; 15:460–7.18. Lee CK, Wu YL, Ding PN, Lord SJ, Inoue A, Zhou C, et al. Impact of specific epidermal growth factor receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus che-motherapy in EGFR-mutant lung cancer: a meta-analysis. J Clin Oncol. 2015; 33:1958–65.19. Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009; 28 Suppl 1:S24–31.20. Hsu WH, Yang JC, Mok TS, Loong HH. Overview of current systemic management of EGFR-mutant NSCLC. Ann Oncol. 2018; 29:i3–9.21. Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017; 376:629–40.22. Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011; 12:735–42.23. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010; 362:2380–8.24. Huang YH, Tseng JS, Hsu KH, Chen KC, Su KY, Yu SL, et al. The impact of different first-line EGFR-TKIs on the clinical outcome of sequential osimertinib treatment in advanced NSCLC with secondary T790M. Sci Rep. 2021; 11:12084.25. Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013; 12:220–9.26. Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015; 16:830–8.27. Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009; 361:958–67.28. U.S. Food and Drug Administrastion. FDA summary of safety and effectiveness data (SSED). Premarket approval (PMA) No. P120019. Cobas EGFR Mutation Test v2. Silver Spring, MD: U.S. Food and Drug Administration;2018.29. Reynolds JP, Tubbs RR, Minca EC, MacNamara S, Almeida FA, Ma PC, et al. EGFR mutational genotyping of liquid based cytology samples obtained via fine needle aspiration (FNA) at endobronchial ultrasound of non-small cell lung cancer (NSCLC). Lung Cancer. 2014; 86:158–63.30. Smouse JH, Cibas ES, Janne PA, Joshi VA, Zou KH, Lindeman NI. EGFR mutations are detected comparably in cyto-logic and surgical pathology specimens of nonsmall cell lung cancer. Cancer. 2009; 117:67–72.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Does the efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor differ according to the type of EGFR mutation in non-small cell lung cancer?
- Analytical and Clinical Validation of a Highly Sensitive NGS-Based ctDNA Assay with Real-World Concordance in Non–Small Cell Lung Cancer
- Rare Mechanism of Acquired Resistance to Osimertinib in Korean Patients with EGFR-mutated Non-small Cell Lung Cancer
- EGFR C797S as a Resistance Mechanism of Lazertinib in Non-small Cell Lung Cancer with EGFR T790M Mutation
- Tissue and Plasma-Based Highly Sensitive Blocker Displacement Amplicon Nanopore Sequencing for EGFR Mutations in Lung Cancer