J Korean Med Sci.
2023 Nov;38(46):e395. 10.3346/jkms.2023.38.e395.
Polygenic Risk Score for Cardiovascular Diseases in Artificial Intelligence Paradigm: A Review
- Affiliations
-
- 1Department of Cardiology, Indraprastha APOLLO Hospitals, New Delhi, India
- 2Asia Pacific Vascular Society, New Delhi, India
- 3Stroke Monitoring and Diagnostic Division, AtheroPoint™, Roseville, CA, USA
- 4Bennett University, Greater Noida, India
- 5School of Bioengineering Sciences and Research, Maharashtra Institute of Technology’s Art, Design and Technology University, Pune, India
- 6Department of Medicine, Division of Cardiology, Queen’s University, Kingston, Canada
- 7Department of Medicine, Division of Cardiology, University of Toronto, Toronto, Canada
- 8Heart and Vascular Institute, Adventist Health St. Helena, St. Helena, CA, USA
- 9Department of Vascular Surgery, Central Clinic of Athens, Athens, Greece
- 10Invasive Cardiology Division, University of Szeged, Szeged, Hungary
- 11Department of Food Science and Technology, Graphic Era Deemed to be University, Dehradun, Uttarakhand, India
- 12Department of Radiology, Harvard Medical School, Boston, MA, USA
- 13Department of Vascular Surgery, University of Lisbon, Lisbon, Portugal
- 14Department of Neurology & Stroke Program, University of Maryland, Baltimore, MD, USA
- 15Vascular Screening and Diagnostic Centre and University of Nicosia Medical School, Cyprus
- 16Nephrology Department, Kaiser Permanente, Sacramento, CA, USA
- 17Ann and Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, USA
- 18Allergy, Clinical Immunology and Rheumatology Institute, Toronto, ON, Canada
- 19Department of Radiobiology and Molecular Genetics, National Institute of The Republic of Serbia, University of Belgrade, Beograd, Serbia
- 20MV Diabetes Centre, Royapuram, Chennai, Tamil Nadu, India
- 21Department of Anaesthesiology, AIIMS, New Delhi, India
- 22Department of Electrical and Computer Engineering, Idaho State University, Pocatello, ID, USA
- 23Department of Radiology, Azienda Ospedaliero Universitaria, Cagliari, Italy
- 24Department of Computer Engineering, Graphic Era Deemed to be University, Dehradun, India
- KMID: 2548544
- DOI: http://doi.org/10.3346/jkms.2023.38.e395
Abstract
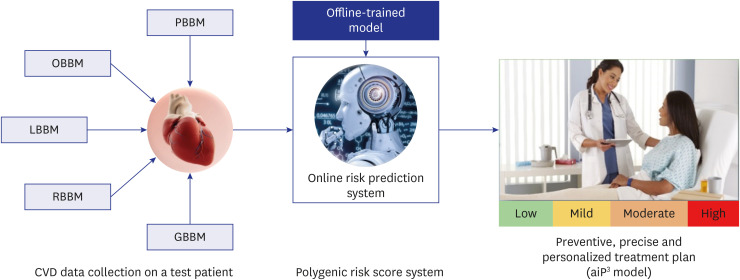
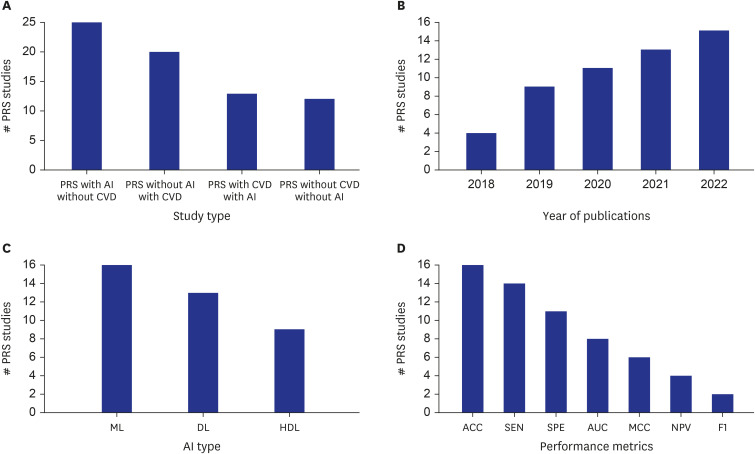
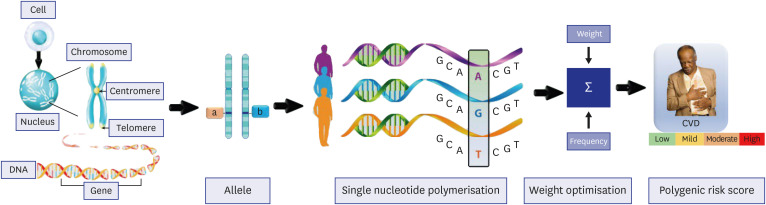
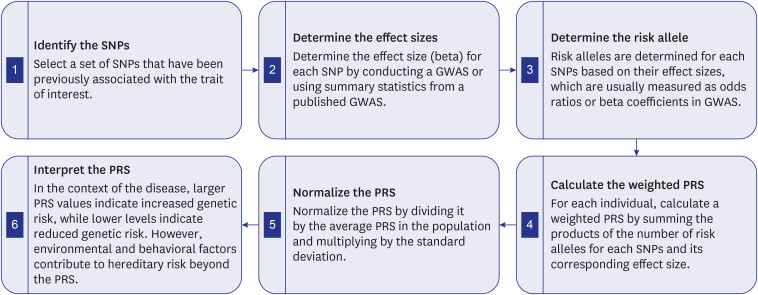
- Cardiovascular disease (CVD) related mortality and morbidity heavily strain society. The relationship between external risk factors and our genetics have not been well established. It is widely acknowledged that environmental influence and individual behaviours play a significant role in CVD vulnerability, leading to the development of polygenic risk scores (PRS). We employed the PRISMA search method to locate pertinent research and literature to extensively review artificial intelligence (AI)-based PRS models for CVD risk prediction. Furthermore, we analyzed and compared conventional vs. AI-based solutions for PRS. We summarized the recent advances in our understanding of the use of AI-based PRS for risk prediction of CVD. Our study proposes three hypotheses: i) Multiple genetic variations and risk factors can be incorporated into AI-based PRS to improve the accuracy of CVD risk predicting. ii) AI-based PRS for CVD circumvents the drawbacks of conventional PRS calculators by incorporating a larger variety of genetic and non-genetic components, allowing for more precise and individualised risk estimations. iii) Using AI approaches, it is possible to significantly reduce the dimensionality of huge genomic datasets, resulting in more accurate and effective disease risk prediction models. Our study highlighted that the AI-PRS model outperformed traditional PRS calculators in predicting CVD risk. Furthermore, using AI-based methods to calculate PRS may increase the precision of risk predictions for CVD and have significant ramifications for individualized prevention and treatment plans.
Keyword
Figure
Reference
-
1. Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, et al. Heart disease and stroke statistics—2020 update: a report from the American Heart Association. Circulation. 2020; 141(9):e139–e596. PMID: 31992061.2. Roth GA, Johnson CO, Abate KH, Abd-Allah F, Ahmed M, Alam K, et al. The burden of cardiovascular diseases among US states, 1990–2016. JAMA Cardiol. 2018; 3(5):375–389. PMID: 29641820.3. O’Donnell CJ, Nabel EG. Genomics of cardiovascular disease. N Engl J Med. 2011; 365(22):2098–2109. PMID: 22129254.
Article4. Gluba A, Banach M, Mikhailidis DP, Rysz J. Genetic determinants of cardiovascular disease: the renin-angiotensin-aldosterone system, paraoxonases, endothelin-1, nitric oxide synthase and adrenergic receptors. In Vivo. 2009; 23(5):797–812. PMID: 19779116.5. Barua JD, Omit SB, Rana HK, Podder NK, Chowdhury UN, Rahman MH. Bioinformatics and system biological approaches for the identification of genetic risk factors in the progression of cardiovascular disease. Cardiovasc Ther. 2022; 2022:9034996. PMID: 36035865.
Article6. Phan JH, Quo CF, Wang MD. Cardiovascular genomics: a biomarker identification pipeline. IEEE Trans Inf Technol Biomed. 2012; 16(5):809–822. PMID: 22614726.
Article7. Maniruzzaman M, Jahanur Rahman M, Ahammed B, Abedin MM, Suri HS, Biswas M, et al. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput Methods Programs Biomed. 2019; 176:173–193. PMID: 31200905.
Article8. Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun. 2020; 11(1):163. PMID: 31919418.9. Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012; 90(1):7–24. PMID: 22243964.
Article10. Gallagher MD, Chen-Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genet. 2018; 102(5):717–730. PMID: 29727686.
Article11. Nolte IM, Munoz ML, Tragante V, Amare AT, Jansen R, Vaez A, et al. Genetic loci associated with heart rate variability and their effects on cardiac disease risk. Nat Commun. 2017; 8(1):15805. PMID: 28613276.12. Wu JH, Lemaitre RN, Manichaikul A, Guan W, Tanaka T, Foy M, et al. Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2013; 6(2):171–183. PMID: 23362303.
Article13. Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010; 376(9750):1393–1400. PMID: 20971364.
Article14. El-Baz AS, Suri JS. Cardiovascular and Coronary Artery Imaging: Volume 1. Cambridge, MA, USA: Academic Press;2021.15. Khanna NN, Maindarkar M, Puvvula A, Paul S, Bhagawati M, Ahluwalia P, et al. Vascular implications of COVID-19: role of radiological imaging, artificial intelligence, and tissue characterization: a special report. J Cardiovasc Dev Dis. 2022; 9(8):268. PMID: 36005433.
Article16. Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009; 302(1):37–48. PMID: 19567438.
Article17. Khanna NN, Maindarkar M, Saxena A, Ahluwalia P, Paul S, Srivastava SK, et al. Cardiovascular/stroke risk assessment in patients with erectile dysfunction-a role of carotid wall arterial imaging and plaque tissue characterization using artificial intelligence paradigm: a narrative review. Diagnostics (Basel). 2022; 12(5):1249. PMID: 35626404.
Article18. Suri JS, Paul S, Maindarkar MA, Puvvula A, Saxena S, Saba L, et al. Cardiovascular/stroke risk stratification in Parkinson’s disease patients using atherosclerosis pathway and artificial intelligence paradigm: a systematic review. Metabolites. 2022; 12(4):312. PMID: 35448500.
Article19. Cahill TJ, Ashrafian H, Watkins H. Genetic cardiomyopathies causing heart failure. Circ Res. 2013; 113(6):660–675. PMID: 23989711.
Article20. Hucker WJ, Saini H, Lubitz SA, Ellinor PT. Atrial fibrillation genetics: is there a practical clinical value now or in the future? Can J Cardiol. 2016; 32(11):1300–1305. PMID: 27094126.
Article21. Weiss JC, Natarajan S, Peissig PL, McCarty CA, Page D. Machine learning for personalized medicine: predicting primary myocardial infarction from electronic health records. AI Mag. 2012; 33(4):33–33.
Article22. Shameer K, Johnson KW, Glicksberg BS, Dudley JT, Sengupta PP. Machine learning in cardiovascular medicine: are we there yet? Heart. 2018; 104(14):1156–1164. PMID: 29352006.23. Kagiyama N, Shrestha S, Farjo PD, Sengupta PP. Artificial intelligence: practical primer for clinical research in cardiovascular disease. J Am Heart Assoc. 2019; 8(17):e012788. PMID: 31450991.
Article24. Alimadadi A, Manandhar I, Aryal S, Munroe PB, Joe B, Cheng X. Machine learning-based classification and diagnosis of clinical cardiomyopathies. Physiol Genomics. 2020; 52(9):391–400. PMID: 32744882.25. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In : Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, 18th International Conference; October 5-9, 2015; Munich, Germany. Berlin, Germany: Springer;2015. p. 234–241.26. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In : Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; June 7-12, 2015; Boston, MA, USA. Washington, D.C., USA: IEEE Computer Society;2015. p. 3431–3440.27. Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017; 39(12):2481–2495. PMID: 28060704.
Article28. Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. In : Proceedings of the IEEE International Conference on Computer Vision; December 7-13, 2015; Santiago, Chile. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2015. p. 1520–1528.29. Karim F, Majumdar S, Darabi H, Harford S. Multivariate LSTM-FCNs for time series classification. Neural Netw. 2019; 116:237–245. PMID: 31121421.
Article30. Xia M, Yan W, Huang Y, Guo Y, Zhou G, Wang Y. Extracting membrane borders in IVUS images using a multi-scale feature aggregated u-net. 2020. In : 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC); July 20-24, 2020; Montreal, Canada. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2020. p. 1650–1653.31. Azad R, Asadi-Aghbolaghi M, Fathy M, Escalera S. Bi-directional ConvLSTM U-Net with densley connected convolutions. In : Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops; October 27-28, 2019; Seoul, Korea. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2019. p. 406–415.32. Wollmann T, Gunkel M, Chung I, Erfle H, Rippe K, Rohr K. GRUU-Net: Integrated convolutional and gated recurrent neural network for cell segmentation. Med Image Anal. 2019; 56:68–79. PMID: 31200289.
Article33. Adak A, Pradhan B, Shukla N, Alamri A. Unboxing deep learning model of food delivery service reviews using explainable artificial intelligence (XAI) technique. Foods. 2022; 11(14):2019. PMID: 35885262.
Article34. Deif MA, Solyman AA, Kamarposhti MA, Band SS, Hammam RE. A deep bidirectional recurrent neural network for identification of SARS-CoV-2 from viral genome sequences. Math Biosci Eng. 2021; 18(6):8933–8950. PMID: 34814329.
Article35. Suri JS, Bhagawati M, Agarwal S, Paul S, Pandey A, Gupta SK, et al. UNet deep learning architecture for segmentation of vascular and non-vascular images: a microscopic look at UNet components buffered with pruning, explainable artificial intelligence, and bias. IEEE Access. 2022; 11:595–645.
Article36. Libiseller-Egger J, Phelan JE, Attia ZI, Benavente ED, Campino S, Friedman PA, et al. Deep learning-derived cardiovascular age shares a genetic basis with other cardiac phenotypes. Sci Rep. 2022; 12(1):22625. PMID: 36587059.
Article37. Johri AM, Mantella LE, Jamthikar AD, Saba L, Laird JR, Suri JS. Role of artificial intelligence in cardiovascular risk prediction and outcomes: comparison of machine-learning and conventional statistical approaches for the analysis of carotid ultrasound features and intra-plaque neovascularization. Int J Cardiovasc Imaging. 2021; 37(11):3145–3156. PMID: 34050838.
Article38. Krittanawong C, Johnson KW, Choi E, Kaplin S, Venner E, Murugan M, et al. Artificial intelligence and cardiovascular genetics. Life (Basel). 2022; 12(2):279. PMID: 35207566.
Article39. El-Baz A, Suri JS. Big Data in Multimodal Medical Imaging. Boca Raton, FL, USA: CRC Press;2019.40. Jamthikar AD, Gupta D, Mantella LE, Saba L, Laird JR, Johri AM, et al. Multiclass machine learning vs. conventional calculators for stroke/CVD risk assessment using carotid plaque predictors with coronary angiography scores as gold standard: a 500 participants study. Int J Cardiovasc Imaging. 2021; 37(4):1171–1187. PMID: 33184741.
Article41. Jamthikar A, Gupta D, Khanna NN, Saba L, Laird JR, Suri JS. Cardiovascular/stroke risk prevention: a new machine learning framework integrating carotid ultrasound image-based phenotypes and its harmonics with conventional risk factors. Indian Heart J. 2020; 72(4):258–264. PMID: 32861380.
Article42. Steinfeldt J, Buergel T, Loock L, Kittner P, Ruyoga G, Zu Belzen JU, et al. Neural network-based integration of polygenic and clinical information: development and validation of a prediction model for 10-year risk of major adverse cardiac events in the UK Biobank cohort. Lancet Digit Health. 2022; 4(2):e84–e94. PMID: 35090679.
Article43. Johri AM, Singh KV, Mantella LE, Saba L, Sharma A, Laird JR, et al. Deep learning artificial intelligence framework for multiclass coronary artery disease prediction using combination of conventional risk factors, carotid ultrasound, and intraplaque neovascularization. Comput Biol Med. 2022; 150:106018. PMID: 36174330.
Article44. Konstantonis G, Singh KV, Sfikakis PP, Jamthikar AD, Kitas GD, Gupta SK, et al. Cardiovascular disease detection using machine learning and carotid/femoral arterial imaging frameworks in rheumatoid arthritis patients. Rheumatol Int. 2022; 42(2):215–239. PMID: 35013839.
Article45. Jamthikar A, Gupta D, Johri AM, Mantella LE, Saba L, Suri JS. A machine learning framework for risk prediction of multi-label cardiovascular events based on focused carotid plaque B-Mode ultrasound: a Canadian study. Comput Biol Med. 2022; 140:105102. PMID: 34973521.
Article46. Ho DS, Schierding W, Wake M, Saffery R, O’Sullivan J. Machine learning SNP based prediction for precision medicine. Front Genet. 2019; 10:267. PMID: 30972108.
Article47. O’Sullivan JW, Raghavan S, Marquez-Luna C, Luzum JA, Damrauer SM, Ashley EA, et al. Polygenic risk scores for cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2022; 146(8):e93–118. PMID: 35862132.
Article48. Kuanr M, Mohapatra P, Mittal S, Maindarkar M, Fauda MM, Saba L, et al. Recommender system for the efficient treatment of COVID-19 using a convolutional neural network model and image similarity. Diagnostics (Basel). 2022; 12(11):2700. PMID: 36359545.
Article49. Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017; 135(22):2091–2101. PMID: 28223407.
Article50. Quazi S. Artificial intelligence and machine learning in precision and genomic medicine. Med Oncol. 2022; 39(8):120. PMID: 35704152.
Article51. Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J Am Coll Cardiol. 2018; 72(16):1883–1893. PMID: 30309464.52. Fritzsche MC, Akyüz K, Cano Abadía M, McLennan S, Marttinen P, Mayrhofer MT, et al. Ethical layering in AI-driven polygenic risk scores: new complexities, new challenges. Front Genet. 2023; 14:1098439. PMID: 36816027.
Article53. Aragam KG, Natarajan P. Polygenic scores to assess atherosclerotic cardiovascular disease risk: clinical perspectives and basic implications. Circ Res. 2020; 126(9):1159–1177. PMID: 32324503.
Article54. Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013; 9(3):e1003348. PMID: 23555274.
Article55. Dubey AK, Chabert GL, Carriero A, Pasche A, Danna PS, Agarwal S, et al. Ensemble deep learning derived from transfer learning for classification of COVID-19 patients on hybrid deep-learning-based lung segmentation: a data augmentation and balancing framework. Diagnostics (Basel). 2023; 13(11):1954. PMID: 37296806.
Article56. Suri JS, Agarwal S, Saba L, Chabert GL, Carriero A, Paschè A, et al. Multicenter study on COVID-19 lung computed tomography segmentation with varying glass ground opacities using unseen deep learning artificial intelligence paradigms: COVLIAS 1.0 validation. J Med Syst. 2022; 46(10):62. PMID: 35988110.
Article57. Suri JS, Agarwal S, Chabert GL, Carriero A, Paschè A, Danna PS, et al. COVLIAS 2.0-cXAI: cloud-based explainable deep learning system for COVID-19 lesion localization in computed tomography scans. Diagnostics (Basel). 2022; 12(6):1482. PMID: 35741292.
Article58. Das S, Nayak GK, Saba L, Kalra M, Suri JS, Saxena S. An artificial intelligence framework and its bias for brain tumor segmentation: a narrative review. Comput Biol Med. 2022; 143:105273. PMID: 35228172.
Article59. Suri JS, Agarwal S, Carriero A, Paschè A, Danna PS, Columbu M, et al. COVLIAS 1.0 vs. MedSeg: artificial intelligence-based comparative study for automated COVID-19 computed tomography lung segmentation in Italian and Croatian cohorts. Diagnostics (Basel). 2021; 11(12):2367. PMID: 34943603.
Article60. Suri JS, Agarwal S, Elavarthi P, Pathak R, Ketireddy V, Columbu M, et al. Inter-variability study of COVLIAS 1.0: hybrid deep learning models for COVID-19 lung segmentation in computed tomography. Diagnostics (Basel). 2021; 11(11):2025. PMID: 34829372.
Article61. Jena B, Saxena S, Nayak GK, Saba L, Sharma N, Suri JS. Artificial intelligence-based hybrid deep learning models for image classification: the first narrative review. Comput Biol Med. 2021; 137:104803. PMID: 34536856.
Article62. Jain PK, Sharma N, Giannopoulos AA, Saba L, Nicolaides A, Suri JS. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput Biol Med. 2021; 136:104721. PMID: 34371320.
Article63. Skandha SS, Nicolaides A, Gupta SK, et al. A hybrid deep learning paradigm for carotid plaque tissue characterization and its validation in multicenter cohorts using a supercomputer framework. Comput Biol Med. 2022; 141:105131. PMID: 34922173.
Article64. Zheng H, Wang H, Azuaje F. Incorporation of ontology-driven biological knowledge into cardiovascular genomics. In : 2011 Computing in Cardiology; September 18-21, 2011; Hangzhou, China. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2011. p. 565–568.65. Wung SF, Hickey KT, Taylor JY, Gallek MJ. Cardiovascular genomics. J Nurs Scholarsh. 2013; 45(1):60–68. PMID: 23368089.
Article66. Young WJ, Ramírez J, van Duijvenboden S, et al. Will genetic data significantly change cardiovascular risk prediction in daily practice? In : 2020 Computing in Cardiology; September 13-16, 2020; Rimini, Italy. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2020. p. 1–4.67. Ganesh SK, Arnett DK, Assimes TL, Basson CT, Chakravarti A, Ellinor PT, et al. Genetics and genomics for the prevention and treatment of cardiovascular disease: update: a scientific statement from the American Heart Association. Circulation. 2013; 128(25):2813–2851. PMID: 24297835.
Article68. Tandel GS, Tiwari A, Kakde OG, Gupta N, Saba L, Suri JS. Role of ensemble deep learning for brain tumor classification in multiple magnetic resonance imaging sequence Data. Diagnostics (Basel). 2023; 13(3):481. PMID: 36766587.
Article69. Terrada O, Cherradi B, Raihani A, Bouattane O. Classification and prediction of atherosclerosis diseases using machine learning algorithms. 2019 5th International Conference on Optimization and Applications (ICOA). Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2019. p. 10056.70. Suri JS, Bhagawati M, Paul S, Protogeron A, Sfikakis PP, Kitas GD, et al. Understanding the bias in machine learning systems for cardiovascular disease risk assessment: The first of its kind review. Comput Biol Med. 2022; 142:105204. PMID: 35033879.
Article71. Jamthikar AD, Gupta D, Mantella LE, et al. Multiclass machine learning vs. conventional calculators for stroke/CVD risk assessment using carotid plaque predictors with coronary angiography scores as gold standard: a 500 participants study. Int J Cardiovasc Imaging. 2021; 37(4):1171–1187. PMID: 33184741.
Article72. Reel PS, Reel S, Pearson E, Trucco E, Jefferson E. Using machine learning approaches for multi-omics data analysis: a review. Biotechnol Adv. 2021; 49:107739. PMID: 33794304.
Article73. Vakili D, Radenkovic D, Chawla S, Bhatt DL. Panomics: new databases for advancing cardiology. Front Cardiovasc Med. 2021; 8:587768. PMID: 34041278.
Article74. Picard M, Scott-Boyer MP, Bodein A, Périn O, Droit A. Integration strategies of multi-omics data for machine learning analysis. Comput Struct Biotechnol J. 2021; 19:3735–3746. PMID: 34285775.
Article75. Pan Y, Lei X, Zhang Y. Association predictions of genomics, proteinomics, transcriptomics, microbiome, metabolomics, pathomics, radiomics, drug, symptoms, environment factor, and disease networks: a comprehensive approach. Med Res Rev. 2022; 42(1):441–461. PMID: 34346083.
Article76. Hamamoto R, Komatsu M, Takasawa K, Asada K, Kaneko S. Epigenetics analysis and integrated analysis of multiomics data, including epigenetic data, using artificial intelligence in the era of precision medicine. Biomolecules. 2019; 10(1):62. PMID: 31905969.
Article77. Schnabel RB, Baccarelli A, Lin H, Ellinor PT, Benjamin EJ. Next steps in cardiovascular disease genomic research--sequencing, epigenetics, and transcriptomics. Clin Chem. 2012; 58(1):113–126. PMID: 22100807.
Article78. Jacinto FV, Link W, Ferreira BI. CRISPR/Cas9-mediated genome editing: From basic research to translational medicine. J Cell Mol Med. 2020; 24(7):3766–3778. PMID: 32096600.
Article79. Wang Z, Emmerich A, Pillon NJ, Moore T, Hemerich D, Cornelis MC, et al. Genome-wide association analyses of physical activity and sedentary behavior provide insights into underlying mechanisms and roles in disease prevention. Nat Genet. 2022; 54(9):1332–1344. PMID: 36071172.80. Tahir UA, Katz DH, Avila-Pachecho J, Bick AG, Pampana A, Robbins JM, et al. Whole genome association study of the plasma metabolome identifies metabolites linked to cardiometabolic disease in black individuals. Nat Commun. 2022; 13(1):4923. PMID: 35995766.81. Marees AT, de Kluiver H, Stringer S, Vorspan F, Curis E, Marie-Claire C, et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res. 2018; 27(2):e1608. PMID: 29484742.
Article82. Katz DH, Tahir UA, Bick AG, Pampana A, Ngo D, Benson MD, et al. Whole genome sequence analysis of the plasma proteome in black adults provides novel insights into cardiovascular disease. Circulation. 2022; 145(5):357–370. PMID: 34814699.83. Rahman MH, Peng S, Hu X, Chen C, Rahman MR, Uddin S, et al. A network-based bioinformatics approach to identify molecular biomarkers for type 2 diabetes that are linked to the progression of neurological diseases. Int J Environ Res Public Health. 2020; 17(3):1035. PMID: 32041280.
Article84. Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019; 104(1):21–34. PMID: 30554720.85. Pjanic M, Miller CL, Wirka R, Kim JB, DiRenzo DM, Quertermous T. Genetics and genomics of coronary artery disease. Curr Cardiol Rep. 2016; 18(10):102. PMID: 27586139.
Article86. Musunuru K, Hershberger RE, Day SM, Klinedinst NJ, Landstrom AP, Parikh VN, et al. Genetic testing for inherited cardiovascular diseases: a scientific statement from the American Heart Association. Circ Genom Precis Med. 2020; 13(4):e000067. PMID: 32698598.
Article87. Miyazawa K, Ito K. Genetic analysis for coronary artery disease toward diverse populations. Front Genet. 2021; 12:766485. PMID: 34880905.
Article88. Kwon OS, Hong M, Kim TH, Hwang I, Shim J, Choi EK, et al. Genome-wide association study-based prediction of atrial fibrillation using artificial intelligence. Open Heart. 2022; 9(1):e001898. PMID: 35086918.
Article89. Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020; 15(9):2759–2772. PMID: 32709988.
Article90. Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016; 17(7):392–406. PMID: 27140283.
Article91. Collister JA, Liu X, Clifton L. Calculating polygenic risk scores (PRS) in UK Biobank: a practical guide for epidemiologists. Front Genet. 2022; 13:818574. PMID: 35251129.
Article92. Wand H, Lambert SA, Tamburro C, Iacocca MA, O’Sullivan JW, Sillari C, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature. 2021; 591(7849):211–219. PMID: 33692554.
Article93. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019; 51(4):584–591. PMID: 30926966.
Article94. Hindy G, Aragam KG, Ng K, Chaffin M, Lotta LA, Baras A, et al. Genome-wide polygenic score, clinical risk factors, and long-term trajectories of coronary artery disease. Arterioscler Thromb Vasc Biol. 2020; 40(11):2738–2746. PMID: 32957805.
Article95. de Marvao A, Dawes TJ, O’Regan DP. Artificial intelligence for cardiac imaging-genetics research. Front Cardiovasc Med. 2020; 6:195. PMID: 32039240.
Article96. Öztornaci RO, Coşgun E, Çolak C, Taşdelen B. Prediction of Polygenic Risk Score by machine learning and deep learning methods in genome-wide association studies. bioRxiv.
Article97. Li L, Huang Y, Han Y, Jiang J. Use of deep learning genomics to discriminate Alzheimer’s disease and healthy controls. In : 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC); November 1-5, 2021; Piscataway, NJ, USA. Institute of Electrical and Electronics Engineers;2021. p. 5788–5791.98. Bhadri K, Karnik N, Dhatrak P. Current advancements in cardiovascular disease management using artificial intelligence and machine learning models: current scenario and challenges. In : 2022 10th International Conference on Emerging Trends in Engineering and Technology-Signal and Information Processing (ICETET-SIP-22); April 29-30, 2022; Nagpur, India. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2022. p. 1–6.99. Dai H, Younis A, Kong JD, Puce L, Jabbour G, Yuan H, et al. Big data in cardiology: State-of-art and future prospects. Front Cardiovasc Med. 2022; 9:844296. PMID: 35433868.
Article100. Dai J, Lv J, Zhu M, Wang Y, Qin N, Ma H, et al. Identification of risk loci and a polygenic risk score for lung cancer: a large-scale prospective cohort study in Chinese populations. Lancet Respir Med. 2019; 7(10):881–891. PMID: 31326317.
Article101. Zekavat SM, Raghu VK, Trinder M, Ye Y, Koyama S, Honigberg MC, et al. Deep learning of the retina enables phenome-and genome-wide analyses of the microvasculature. Circulation. 2022; 145(2):134–150. PMID: 34743558.
Article102. Westerlund AM, Hawe JS, Heinig M, Schunkert H. Risk prediction of cardiovascular events by exploration of molecular data with explainable artificial intelligence. Int J Mol Sci. 2021; 22(19):10291. PMID: 34638627.
Article103. Rukhsar L, Bangyal WH, Ali Khan MS, Ag Ibrahim AA, Nisar K, Rawat DB. Analyzing RNA-seq gene expression data using deep learning approaches for cancer classification. Applied Sciences. 2022; 12(4):1850.
Article104. Mathur P, Srivastava S, Xu X, Mehta JL. Artificial intelligence, machine learning, and cardiovascular disease. Clin Med Insights Cardiol. 2020; 14:1179546820927404. PMID: 32952403.
Article105. Suri JS, Maindarkar MA, Paul S, Ahluwalia P, Bhagawati M, Saba L, et al. Deep learning paradigm for cardiovascular disease/stroke risk stratification in Parkinson’s disease affected by COVID-19: a narrative review. Diagnostics (Basel). 2022; 12(7):1543. PMID: 35885449.
Article106. Suri JS, Bhagawati M, Paul S, Protogerou AD, Sfikakis PP, Kitas GD, et al. A powerful paradigm for cardiovascular risk stratification using multiclass, multi-label, and ensemble-based machine learning paradigms: a narrative review. Diagnostics (Basel). 2022; 12(3):722. PMID: 35328275.
Article107. Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One. 2017; 12(4):e0174944. PMID: 28376093.
Article108. Schiano C, Franzese M, Geraci F, Zanfardino M, Maiello C, Palmieri V, et al. Machine learning and bioinformatics framework integration to potential familial DCM-related markers discovery. Genes (Basel). 2021; 12(12):1946. PMID: 34946895.
Article109. Saba L, Tiwari A, Biswas M, Gupta SK, Godia-Cuadrado E, Chaturvedi A, et al. Wilson’s disease: a new perspective review on its genetics, diagnosis and treatment. Front Biosci (Elite Ed). 2019; 11(1):166–185. PMID: 31136971.110. Liu B, Fang L, Xiong Y, Du Q, Xiang Y, Chen X, et al. A machine learning model based on genetic and traditional cardiovascular risk factors to predict premature coronary artery disease. Front Biosci (Landmark Ed). 2022; 27(7):211. PMID: 35866398.
Article111. Ordikhani M, Saniee Abadeh M, Prugger C, Hassannejad R, Mohammadifard N, Sarrafzadegan N. An evolutionary machine learning algorithm for cardiovascular disease risk prediction. PLoS One. 2022; 17(7):e0271723. PMID: 35901181.
Article112. Kannel WB, McGee D, Gordon T. A general cardiovascular risk profile: the Framingham Study. Am J Cardiol. 1976; 38(1):46–51. PMID: 132862.
Article113. Assmann G, Schulte H. The Prospective Cardiovascular Münster (PROCAM) study: prevalence of hyperlipidemia in persons with hypertension and/or diabetes mellitus and the relationship to coronary heart disease. Am Heart J. 1988; 116(6 Pt 2):1713–1724. PMID: 3202078.
Article114. Woodward M, Brindle P, Tunstall-Pedoe H. SIGN Group on Risk Estimation. Adding social deprivation and family history to cardiovascular risk assessment: the ASSIGN score from the Scottish Heart Health Extended Cohort (SHHEC). Heart. 2007; 93(2):172–176. PMID: 17090561.
Article115. Lakoski SG, Greenland P, Wong ND, Schreiner PJ, Herrington DM, Kronmal RA, et al. Coronary artery calcium scores and risk for cardiovascular events in women classified as “low risk” based on Framingham risk score: the multi-ethnic study of atherosclerosis (MESA). Arch Intern Med. 2007; 167(22):2437–2442. PMID: 18071165.
Article116. Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. JAMA. 2007; 297(6):611–619. PMID: 17299196.
Article117. Hippisley-Cox J, Coupland C, Vinogradova Y, Robson J, Minhas R, Sheikh A, et al. Predicting cardiovascular risk in England and Wales: prospective derivation and validation of QRISK2. BMJ. 2008; 336(7659):1475–1482. PMID: 18573856.
Article118. Hippisley-Cox J, Coupland C, Vinogradova Y, Robson J, May M, Brindle P. Derivation and validation of QRISK, a new cardiovascular disease risk score for the United Kingdom: prospective open cohort study. BMJ. 2007; 335(7611):136. PMID: 17615182.
Article119. Goff DC Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014; 129(25):Suppl 2. S49–S73. PMID: 24222018.120. Conroy RM, Pyörälä K, Fitzgerald AP, Sans S, Menotti A, De Backer G, et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: the SCORE project. Eur Heart J. 2003; 24(11):987–1003. PMID: 12788299.
Article121. Khandelwal M, Kumar Rout R, Umer S, Mallik S, Li A. Multifactorial feature extraction and site prognosis model for protein methylation data. Brief Funct Genomics. 2023; 22(1):20–30. PMID: 36310537.
Article122. Pasha SN, Ramesh D, Mohmmad S, Harshavardhan A. Cardiovascular disease prediction using deep learning techniques. IOP Conf Ser Mater Sci Eng. 2020; 981(2):022006.
Article123. Banchhor SK, Londhe ND, Araki T, Saba L, Radeva P, Laird JR, et al. Wall-based measurement features provides an improved IVUS coronary artery risk assessment when fused with plaque texture-based features during machine learning paradigm. Comput Biol Med. 2017; 91:198–212. PMID: 29100114.
Article124. Araki T, Ikeda N, Shukla D, Jain PK, Londhe ND, Shrivastava VK, et al. PCA-based polling strategy in machine learning framework for coronary artery disease risk assessment in intravascular ultrasound: a link between carotid and coronary grayscale plaque morphology. Comput Methods Programs Biomed. 2016; 128:137–158. PMID: 27040838.
Article125. Khalifa NE, Taha MH, Ali DE, Slowik A, Hassanien AE. Artificial intelligence technique for gene expression by tumor RNA-Seq data: a novel optimized deep learning approach. IEEE Access. 2020; 8:22874–22883.
Article126. Peng J, Xue H, Wei Z, Tuncali I, Hao J, Shang X. Integrating multi-network topology for gene function prediction using deep neural networks. Brief Bioinform. 2021; 22(2):2096–2105. PMID: 32249297.
Article127. Jamthikar A, Gupta D, Khanna NN, Saba L, Araki T, Viskovic K, et al. A low-cost machine learning-based cardiovascular/stroke risk assessment system: integration of conventional factors with image phenotypes. Cardiovasc Diagn Ther. 2019; 9(5):420–430. PMID: 31737514.
Article128. Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Methods Programs Biomed. 2017; 150:9–22. PMID: 28859832.
Article129. Araki T, Ikeda N, Dey N, Chakraborty S, Saba L, Kumar D, et al. A comparative approach of four different image registration techniques for quantitative assessment of coronary artery calcium lesions using intravascular ultrasound. Comput Methods Programs Biomed. 2015; 118(2):158–172. PMID: 25523233.
Article130. Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. Reliable and accurate psoriasis disease classification in dermatology images using comprehensive feature space in machine learning paradigm. Expert Syst Appl. 2015; 42(15-16):6184–6195.
Article131. Agarwal M, Agarwal S, Saba L, Chabert GL, Gupta S, Carriero A, et al. Eight pruning deep learning models for low storage and high-speed COVID-19 computed tomography lung segmentation and heatmap-based lesion localization: a multicenter study using COVLIAS 2.0. Comput Biol Med. 2022; 146:105571. PMID: 35751196.
Article132. Song S, Jiang W, Hou L, Zhao H. Leveraging effect size distributions to improve polygenic risk scores derived from summary statistics of genome-wide association studies. PLOS Comput Biol. 2020; 16(2):e1007565. PMID: 32045423.
Article133. Xu Y, Wang Y, Xie X, Wang F, Chen Q, Sun H. An autoencoder-based matrix factorization approach to estimating cell proportion from bulk tumor RNA-seq data. In : 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); December 9-12, 2021; Piscataway, NJ, USA. Institute of Electrical and Electronics Engineers;2021. p. 562–567.134. Zeng M, Lu C, Fei Z, Wu FX, Li Y, Wang J, et al. DMFLDA: a deep learning framework for predicting lncRNA–disease associations. IEEE/ACM Trans Comput Biol Bioinformatics. 2021; 18(6):2353–2363.
Article135. Zhao M, Tang Y, Kim H, Hasegawa K. Machine learning with k-means dimensional reduction for predicting survival outcomes in patients with breast cancer. Cancer Inform. 2018; 17:1176935118810215. PMID: 30455569.
Article136. Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000; 16(10):906–914. PMID: 11120680.
Article137. Gu Y, Zheng S, Yin Q, Jiang R, Li J. REDDA: Integrating multiple biological relations to heterogeneous graph neural network for drug-disease association prediction. Comput Biol Med. 2022; 150:106127. PMID: 36182762.
Article138. Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015; 97(4):576–592. PMID: 26430803.139. Privé F, Aschard H, Blum MG. Efficient implementation of penalized regression for genetic risk prediction. Genetics. 2019; 212(1):65–74. PMID: 30808621.
Article140. Leonenko G, Sims R, Shoai M, Frizzati A, Bossù P, Spalletta G, et al. Polygenic risk and hazard scores for Alzheimer’s disease prediction. Ann Clin Transl Neurol. 2019; 6(3):456–465. PMID: 30911569.
Article141. Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019; 8(7):giz082. PMID: 31307061.
Article142. Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019; 10(1):1776. PMID: 30992449.
Article143. Song L, Horvath S. Predicting COPD status with a random generalized linear model. Syst Biomed (Austin). 2013; 1(4):261–267.
Article144. Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, et al. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation. 2019; 139(13):1593–1602. PMID: 30586733.
Article145. Lloyd-Jones LR, Zeng J, Sidorenko J, Yengo L, Moser G, Kemper KE, et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat Commun. 2019; 10(1):5086. PMID: 31704910.
Article146. Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018; 50(11):1505–1513. PMID: 30297969.
Article147. Munjral S, Maindarkar M, Ahluwalia P, Puvvula A, Jamthikar A, Jujaray T, et al. Cardiovascular risk stratification in diabetic retinopathy via atherosclerotic pathway in COVID-19/non-COVID-19 frameworks using artificial intelligence paradigm: a narrative review. Diagnostics (Basel). 2022; 12(5):1234. PMID: 35626389.
Article148. Tan CH, Bonham LW, Fan CC, Mormino EC, Sugrue LP, Broce IJ, et al. Polygenic hazard score, amyloid deposition and Alzheimer’s neurodegeneration. Brain. 2019; 142(2):460–470. PMID: 30689776.
Article149. Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol Pharm. 2016; 13(5):1445–1454. PMID: 27007977.
Article150. Kavitha MS, Gangadaran P, Jackson A, Venmathi Maran BA, Kurita T, Ahn BC. Deep neural network models for colon cancer screening. Cancers (Basel). 2022; 14(15):3707. PMID: 35954370.
Article151. Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FC, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018; 362(6420):eaat8464. PMID: 30545857.152. Vlachopoulos C, Aznaouridis K, Ioakeimidis N, Rokkas K, Vasiliadou C, Alexopoulos N, et al. Unfavourable endothelial and inflammatory state in erectile dysfunction patients with or without coronary artery disease. Eur Heart J. 2006; 27(22):2640–2648. PMID: 17056702.
Article153. Gandaglia G, Briganti A, Jackson G, Kloner RA, Montorsi F, Montorsi P, et al. A systematic review of the association between erectile dysfunction and cardiovascular disease. Eur Urol. 2014; 65(5):968–978. PMID: 24011423.
Article154. Suri JS, Agarwal S, Gupta S, Puvvula A, Viskovic K, Suri N, et al. Systematic review of artificial intelligence in acute respiratory distress syndrome for COVID-19 lung patients: a biomedical imaging perspective. IEEE J Biomed Health Inform. 2021; 25(11):4128–4139. PMID: 34379599.
Article155. Paul S, Maindarkar M, Saxena S, Saba L, Turk M, Kalra M, et al. Bias investigation in artificial intelligence systems for early detection of Parkinson’s disease: a narrative review. Diagnostics (Basel). 2022; 12(1):166. PMID: 35054333.
Article156. Suri JS, Agarwal S, Jena B, Saxena S, El-Baz A, Agarwal V, et al. Five strategies for bias estimation in artificial intelligence-based hybrid deep learning for acute respiratory distress syndrome COVID-19 lung infected patients using AP(ai)Bias 2.0: a systematic review. IEEE Trans Instrum Meas. 2022.
Article157. Kariuki JK, Stuart-Shor EM, Leveille SG, Hayman LL. Evaluation of the performance of existing non-laboratory based cardiovascular risk assessment algorithms. BMC Cardiovasc Disord. 2013; 13:123. PMID: 24373202.
Article158. Slack D, Hilgard S, Jia E, Singh S, Lakkaraju H. Fooling LIME and SHAP: adversarial attacks on post hoc explanation methods. Proceedings of the AAAI/ACM Conference on AI, Ethics, and Society. New York, NY, USA: Association for Computing Machinery;2020. p. 180–186.159. Biswas M, Kuppili V, Saba L, Edla DR, Suri HS, Cuadrado-Godia E, et al. State-of-the-art review on deep learning in medical imaging. Front Biosci (Landmark Ed). 2019; 24(3):392–426. PMID: 30468663.
Article160. Jena B, Saxena S, Nayak GK, Balestrieri A, Gupta N, Khanna NN, et al. Brain tumor characterization using radiogenomics in artificial intelligence framework. Cancers (Basel). 2022; 14(16):4052. PMID: 36011048.
Article161. Sanagala SS, Nicolaides A, Gupta SK, Koppula VK, Saba L, Agarwal S, et al. Ten fast transfer learning models for carotid ultrasound plaque tissue characterization in augmentation framework embedded with heatmaps for stroke risk stratification. Diagnostics (Basel). 2021; 11(11):2109. PMID: 34829456.
Article162. Khanna NN, Maindarkar MA, Viswanathan V, Fernandes JF, Paul S, Bhagawati M, et al. Economics of artificial intelligence in healthcare: diagnosis vs. treatment. Healthcare (Basel). 2022; 10(12):2493. PMID: 36554017.
Article163. Pennisi M, Kavasidis I, Spampinato C, Schinina V, Palazzo S, Salanitri FP, et al. An explainable AI system for automated COVID-19 assessment and lesion categorization from CT-scans. Artif Intell Med. 2021; 118:102114. PMID: 34412837.
Article164. Langlotz CP, Allen B, Erickson BJ, Kalpathy-Cramer J, Bigelow K, Cook TS, et al. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology. 2019; 291(3):781–791. PMID: 30990384.
Article165. Collin CB, Gebhardt T, Golebiewski M, Karaderi T, Hillemanns M, Khan FM, et al. Computational models for clinical applications in personalized medicine-guidelines and recommendations for data integration and model validation. J Pers Med. 2022; 12(2):166. PMID: 35207655.
Article166. Khanna NN, Maindarkar MA, Viswanathan V, Puvvula A, Paul S, Bhagawati M, et al. Cardiovascular/stroke risk stratification in diabetic foot infection patients using deep learning-based artificial intelligence: an investigative study. J Clin Med. 2022; 11(22):6844. PMID: 36431321.
Article167. Haque AK, Arifuzzaman BM, Siddik SA, Kalam A, Shahjahan TS, Saleena TS, et al. Semantic web in healthcare: a systematic literature review of application, research gap, and future research avenues. Int J Clin Pract. 2022; 2022:6807484. PMID: 36320897.
Article168. Panwar A, Semwal G, Goel S, Gupta S. Stratification of the lesions in color fundus images of diabetic retinopathy patients using deep learning models and machine learning classifiers. Patgiri R, Bandyopadhyay S, Borah MD, Emilia Balas V, editors. Edge Analytics. Lecture Notes in Electrical Engineering. Singapore: Springer;2022. p. 653–666.169. Garg I, Panda P, Roy K. A low effort approach to structured CNN design using PCA. IEEE Access. 2019; 8:1347–1360.
Article170. Acharya UR, Mookiah MR, Vinitha Sree S, Yanti R, Martis RJ, Saba L, et al. Evolutionary algorithm-based classifier parameter tuning for automatic ovarian cancer tissue characterization and classification. Ultraschall Med. 2014; 35(3):237–245. PMID: 23258769.
Article171. Xuan J, Jiang H, Hu Y, Ren Z, Zou W, Luo Z, et al. Towards effective bug triage with software data reduction techniques. IEEE Trans Knowl Data Eng. 2014; 27(1):264–280.
Article172. Shui L, Ren H, Yang X, Li J, Chen Z, Yi C, et al. The era of radiogenomics in precision medicine: an emerging approach to support diagnosis, treatment decisions, and prognostication in oncology. Front Oncol. 2021; 10:570465. PMID: 33575207.
Article173. Panayides AS, Pattichis MS, Leandrou S, Pitris C, Constantinidou A, Pattichis CS. Radiogenomics for precision medicine with a big data analytics perspective. IEEE J Biomed Health Inform. 2019; 23(5):2063–2079. PMID: 30596591.
Article174. Liu Z, Keller PJ. Emerging imaging and genomic tools for developmental systems biology. Dev Cell. 2016; 36(6):597–610. PMID: 27003934.
Article175. Abdel Razek AA, Alksas A, Shehata M, AbdelKhalek A, Abdel Baky K, El-Baz A, et al. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging. Insights Imaging. 2021; 12(1):152. PMID: 34676470.
Article176. Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S. Emerging applications of artificial intelligence in neuro-oncology. Radiology. 2019; 290(3):607–618. PMID: 30667332.
Article177. Gu X, Yu X, Shi G, Li Y, Yang L. Can PD-L1 expression be predicted by contrast-enhanced CT in patients with gastric adenocarcinoma? A preliminary retrospective study. Abdom Radiol (NY). 2023; 48(1):220–228. PMID: 36271155.
Article178. Srivastava SK, Singh SK, Suri JS. Effect of incremental feature enrichment on healthcare text classification system: a machine learning paradigm. Comput Methods Programs Biomed. 2019; 172:35–51. PMID: 30902126.
Article179. Banchhor SK, Londhe ND, Araki T, Saba L, Radeva P, Laird JR, et al. Well-balanced system for coronary calcium detection and volume measurement in a low resolution intravascular ultrasound videos. Comput Biol Med. 2017; 84:168–181. PMID: 28390284.
Article180. Khalil RA, Saeed N, Masood M, Fard YM, Alouini MS, Al-Naffouri TY. 2 Deep learning in the industrial internet of things: Potentials, challenges, and emerging applications. IEEE Internet Things J. 2021; 8(14):11016–11040.
Article181. Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: a first comparative study of its kind. Comput Methods Programs Biomed. 2016; 126:98–109. PMID: 26830378.
Article182. Karniadakis GE, Kevrekidis IG, Lu L, Perdikaris P, Wang S, Yang L. Physics-informed machine learning. Nat Rev Phys. 2021; 3(6):422–440.
Article183. Biswas M, Kuppili V, Edla DR, Suri HS, Saba L, Marinhoe RT, et al. Symtosis: a liver ultrasound tissue characterization and risk stratification in optimized deep learning paradigm. Comput Methods Programs Biomed. 2018; 155:165–177. PMID: 29512496.
Article184. Roslan RB, Razly IN, Sabri N, Ibrahim Z. Evaluation of psoriasis skin disease classification using convolutional neural network. IAES Int J Artif Intell. 2020; 9(2):349.
Article185. Jamthikar AD, Gupta D, Saba L, Khanna NN, Viskovic K, Mavrogeni S, et al. Artificial intelligence framework for predictive cardiovascular and stroke risk assessment models: a narrative review of integrated approaches using carotid ultrasound. Comput Biol Med. 2020; 126:104043. PMID: 33065389.
Article186. Jamthikar AD, Gupta D, Johri AM, Mantella LE, Saba L, Kolluri R, et al. Low-cost office-based cardiovascular risk stratification using machine learning and focused carotid ultrasound in an Asian-Indian cohort. J Med Syst. 2020; 44(12):208. PMID: 33175247.
Article187. Jamthikar A, Gupta D, Saba L, Khanna NN, Araki T, Viskovic K, et al. Cardiovascular/stroke risk predictive calculators: a comparison between statistical and machine learning models. Cardiovasc Diagn Ther. 2020; 10(4):919–938. PMID: 32968651.
Article188. Bartels S, Franco AR, Rundek T. Carotid intima-media thickness (cIMT) and plaque from risk assessment and clinical use to genetic discoveries. Perspect Med. 2012; 1(1-12):139–145.
Article189. Suri JS, Puvvula A, Biswas M, Majhail M, Saba L, Faa G, et al. COVID-19 pathways for brain and heart injury in comorbidity patients: a role of medical imaging and artificial intelligence-based COVID severity classification: a review. Comput Biol Med. 2020; 124:103960. PMID: 32919186.
Article190. Liu K, Suri JS. Automatic Vessel Indentification for Angiographic Screening. Patent No.: US6845260B2. Alexandria, VA, USA: U.S. Patent and Trademark Office;2005.191. El-Baz A, Gimel’farb G, Suri JS. Stochastic Modeling for Medical Image Analysis. Boca Raton, FL, USA: CRC Press;2015.192. Gupta N, Gupta SK, Pathak RK, Jain V, Rashidi P, Suri JS. Human activity recognition in artificial intelligence framework: a narrative review. Artif Intell Rev. 2022; 55(6):4755–4808. PMID: 35068651.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Artificial Intelligence in Pathology
- Role of artificial intelligence in diagnosing Barrett’s esophagus-related neoplasia
- Polygenic Risk Score and Precision Medicine in Diabetes
- Exploring the Applicability of Artificial Intelligence for the Improvement of Nursing Practice in Korea
- Application of Artificial Intelligence to Cardiovascular Computed Tomography