Clin Exp Otorhinolaryngol.
2023 Nov;16(4):342-358. 10.21053/ceo.2023.00668.
A Novel EYA1 Mutation Causing Alternative RNA Splicing in a Chinese Family With Branchio-Oto Syndrome: Implications for Molecular Diagnosis and Clinical Application
- Affiliations
-
- 1Department of Otorhinolaryngology, Xiangya Hospital, Central South University, Changsha, China
- 2Key Laboratory of Otolaryngology Major Disease Research of Hunan Province, Changsha, China
- 3National Clinical Research Centre for Geriatric Disorders, Department of Geriatrics, Xiangya Hospital, Central South University, Changsha, China
- 4Medical Functional Experiment Center, School of Basic Medicine, Central South University, Changsha, China
- 5Department of Pathology, Xiangya Hospital, Central South University, Changsha, China
- 6Department of Otorhinolaryngology, Head and Neck Surgery, The Affiliated Changsha Central Hospital, Hengyang Medical School, University of South China, Changsha, China
- 7MOE Key Lab of Rare Pediatric Diseases and Institute of Otorhinolaryngology, Head and Neck Surgery, University of South China, Changsha, China
- 8The Hengyang Key Laboratory of Cellular Stress Biology, Institute of Cytology and Genetics, Hengyang Medical School, University of South China, Hengyang, China
- 9Department of Otorhinolaryngology, The Affiliated Maternal and Child Health Hospital of Hunan Province, Hengyang Medical School, University of South China, Changsha, China
- KMID: 2548363
- DOI: http://doi.org/10.21053/ceo.2023.00668
Abstract
Objectives
. Branchio-oto syndrome (BOS) primarily manifests as hearing loss, preauricular pits, and branchial defects. EYA1 is the most common pathogenic gene, and splicing mutations account for a substantial proportion of cases. However, few studies have addressed the structural changes in the protein caused by splicing mutations and potential pathogenic factors, and several studies have shown that middle-ear surgery has limited effectiveness in improving hearing in these patients. BOS has also been relatively infrequently reported in the Chinese population. This study explored the genetic etiology in the family of a proband with BOS and provided clinical treatment to improve the patient’s hearing.
Methods
. We collected detailed clinical features and peripheral blood samples from the patients and unaffected individuals within the family. Pathogenic mutations were identified by whole-exome sequencing and cosegregation analysis and classified according to the American College of Medical Genetics and Genomics guidelines. Alternative splicing was verified through a minigene assay. The predicted three-dimensional protein structure and biochemical experiments were used to investigate the pathogenicity of the mutation. The proband underwent middle-ear surgery and was followed up at 1 month and 6 months postoperatively to monitor auditory improvement.
Results
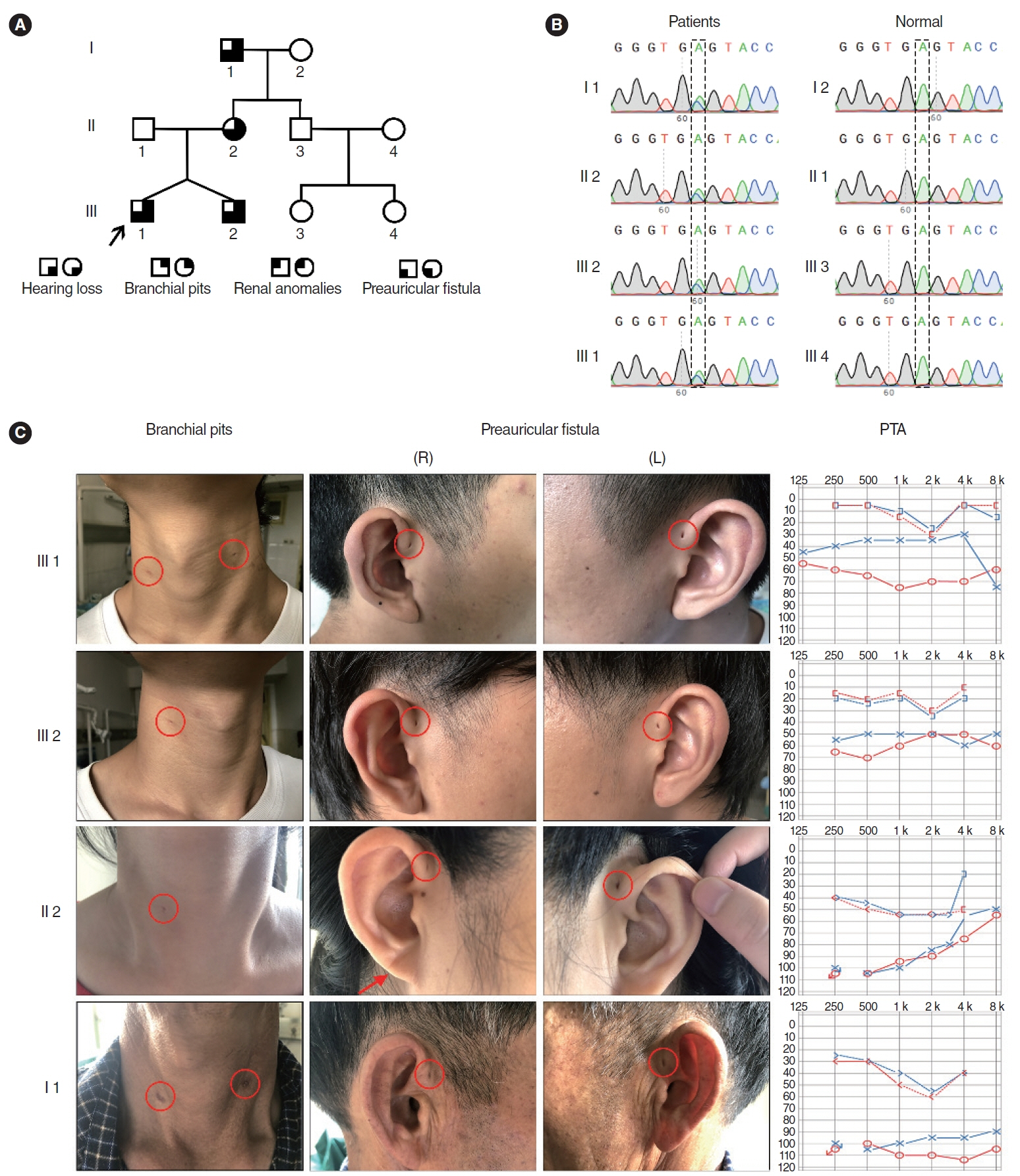
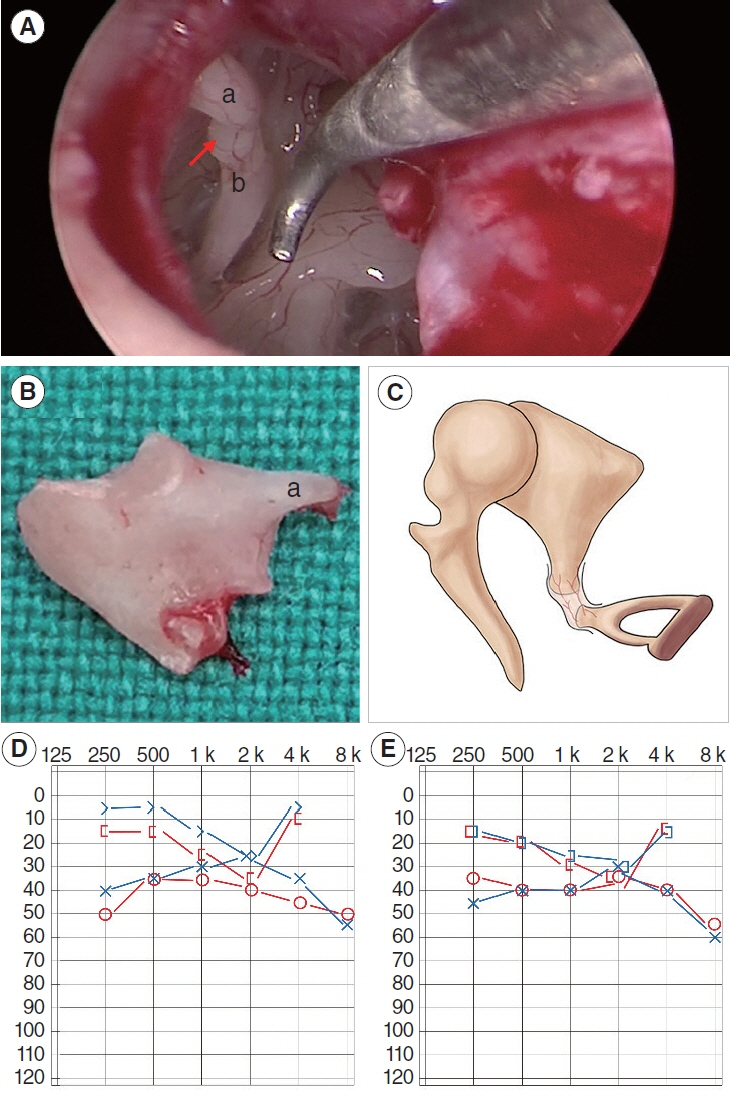
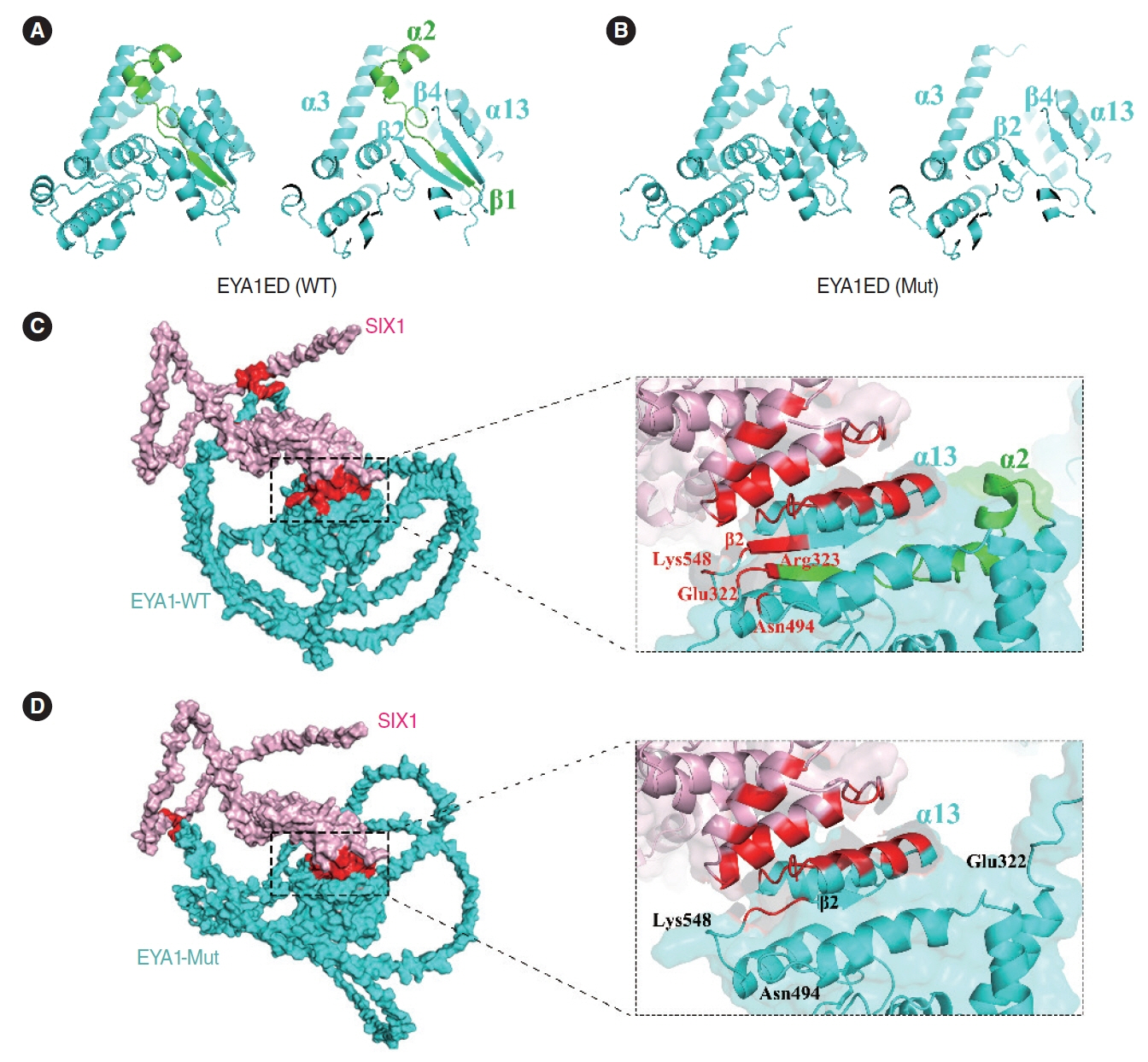
. A novel heterozygous EYA1 splicing variant (c.1050+4 A>C) was identified and classified as pathogenic (PVS1(RNA), PM2, PP1). Skipping of exon 11 of the EYA1 pre-mRNA was confirmed using a minigene assay. This mutation may impair EYA1-SIX1 interactions, as shown by an immunoprecipitation assay. The EYA1-Mut protein exhibited cellular mislocalization and decreased protein expression in cytological experiments. Middle-ear surgery significantly improved hearing loss caused by bone-conduction abnormalities in the proband.
Conclusion
. We reported a novel splicing variant of EYA1 in a Chinese family with BOS and revealed the potential molecular pathogenic mechanism. The significant hearing improvement observed in the proband after middle-ear surgery provides a reference for auditory rehabilitation in similar patients.
Figure
Reference
-
1. Thienpont B, Dimitriadou E, Theodoropoulos K, Breckpot J, Fryssira H, Kitsiou-Tzeli S, et al. Refining the locus of branchio-otic syndrome 2 (BOS2) to a 5.25 Mb locus on chromosome 1q31.3q32.1. Eur J Med Genet. 2009; Nov-Dec. 52(6):393–7.2. Morisada N, Nozu K, Iijima K. Branchio-oto-renal syndrome: comprehensive review based on nationwide surveillance in Japan. Pediatr Int. 2014; Jun. 56(3):309–14.3. Unzaki A, Morisada N, Nozu K, Ye MJ, Ito S, Matsunaga T, et al. Clinically diverse phenotypes and genotypes of patients with branchiooto-renal syndrome. J Hum Genet. 2018; May. 63(5):647–56.4. Fraser FC, Sproule JR, Halal F. Frequency of the branchio-oto-renal (BOR) syndrome in children with profound hearing loss. Am J Med Genet. 1980; 7(3):341–9.5. Kim HR, Song MH, Kim MA, Kim YR, Lee KY, Sonn JK, et al. Identification of a novel nonsynonymous mutation of EYA1 disrupting splice site in a Korean patient with BOR syndrome. Mol Biol Rep. 2014; Jul. 41(7):4321–7.6. Kochhar A, Orten DJ, Sorensen JL, Fischer SM, Cremers CW, Kimberling WJ, et al. SIX1 mutation screening in 247 branchio-oto-renal syndrome families: a recurrent missense mutation associated with BOR. Hum Mutat. 2008; Apr. 29(4):565.7. Zou D, Silvius D, Davenport J, Grifone R, Maire P, Xu PX. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev Biol. 2006; May. 293(2):499–512.8. Moody SA, LaMantia AS. Transcriptional regulation of cranial sensory placode development. Curr Top Dev Biol. 2015; 111:301–50.9. Shah AM, Krohn P, Baxi AB, Tavares AL, Sullivan CH, Chillakuru YR, et al. Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis Model Mech. 2020; Mar. 13(3):dmm043489.10. Chen A, Song J, Acke FR, Mei L, Cai X, Feng Y, et al. Otological manifestations in branchiootorenal spectrum disorder: a systematic review and meta-analysis. Clin Genet. 2021; Jul. 100(1):3–13.11. Song MH, Kwon TJ, Kim HR, Jeon JH, Baek JI, Lee WS, et al. Mutational analysis of EYA1, SIX1 and SIX5 genes and strategies for management of hearing loss in patients with BOR/BO syndrome. PLoS One. 2013; Jun. 8(6):e67236.12. Chang EH, Menezes M, Meyer NC, Cucci RA, Vervoort VS, Schwartz CE, et al. Branchio-oto-renal syndrome: the mutation spectrum in EYA1 and its phenotypic consequences. Hum Mutat. 2004; Jun. 23(6):582–9.13. Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004; May. 101(21):8090–5.14. Gettelfinger JD, Dahl JP. Syndromic hearing loss: a brief review of common presentations and genetics. J Pediatr Genet. 2018; Mar. 7(1):1–8.15. Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997; Feb. 15(2):157–64.16. Propst EJ, Blaser S, Gordon KA, Harrison RV, Papsin BC. Temporal bone findings on computed tomography imaging in branchio-otorenal syndrome. Laryngoscope. 2005; Oct. 115(10):1855–62.17. Feng H, Xu H, Chen B, Sun S, Zhai R, Zeng B, et al. Genetic and phenotypic variability in Chinese patients with branchio-oto-renal or branchio-oto syndrome. Front Genet. 2021; Nov. 12:765433.18. Sang S, Ling J, Liu X, Mei L, Cai X, Li T, et al. Proband whole-exome sequencing identified genes responsible for autosomal recessive nonsyndromic hearing loss in 33 Chinese nuclear families. Front Genet. 2019; Jul. 10:639.19. Li H, Durbin R. Fast and accurate short read alignment with BurrowsWheeler transform. Bioinformatics. 2009; Jul. 25(14):1754–60.20. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; Aug. 25(16):2078–9.21. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; Sep. 38(16):e164.22. Johansson-Akhe I, Wallner B. Improving peptide-protein docking with AlphaFold-Multimer using forced sampling. Front Bioinform. 2022; Sep. 2:959160.23. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019; Jan. 47(D1):D886–94.24. Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002; Mar. 12(3):436–46.25. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; May. 17(5):405–24.26. Walker LC, Hoya M, Wiggins GA, Lindy A, Vincent LM, Parsons MT, et al. Using the ACMG/AMP framework to capture evidence related to predicted and observed impact on splicing: recommendations from the ClinGen SVI Splicing Subgroup. Am J Hum Genet. 2023; Jul. 110(7):1046–67.27. Stockley TL, Mendoza-Londono R, Propst EJ, Sodhi S, Dupuis L, Papsin BC. A recurrent EYA1 mutation causing alternative RNA splicing in branchio-oto-renal syndrome: implications for molecular diagnostics and disease mechanism. Am J Med Genet A. 2009; Mar. 149(3):322–7.28. Orten DJ, Fischer SM, Sorensen JL, Radhakrishna U, Cremers CW, Marres HA, et al. Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum Mutat. 2008; Apr. 29(4):537–44.29. Vervoort VS, Smith RJ, O’Brien J, Schroer R, Abbott A, Stevenson RE, et al. Genomic rearrangements of EYA1 account for a large fraction of families with BOR syndrome. Eur J Hum Genet. 2002; Nov. 10(11):757–66.30. Clarke JC, Honey EM, Bekker E, Snyman LC, Raymond RM Jr, Lord C, et al. A novel nonsense mutation in the EYA1 gene associated with branchio-oto-renal/branchiootic syndrome in an Afrikaner kindred. Clin Genet. 2006; Jul. 70(1):63–7.31. Mann N, Braun DA, Amann K, Tan W, Shril S, Connaughton DM, et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol. 2019; Feb. 30(2):201–15.32. Krug P, Moriniere V, Marlin S, Koubi V, Gabriel HD, Colin E, et al. Mutation screening of the EYA1, SIX1, and SIX5 genes in a large cohort of patients harboring branchio-oto-renal syndrome calls into question the pathogenic role of SIX5 mutations. Hum Mutat. 2011; Feb. 32(2):183–90.33. Kwon MJ, Boo SH, Kim HJ, Cho YS, Chung WH, Hong SH. A novel splice site mutation in the EYA1 gene in a Korean family with branchio-oto (BO) syndrome. Acta Otolaryngol. 2009; Jun. 129(6):688–93.34. Chen P, Liu H, Lin Y, Xu J, Zhu W, Wu H, et al. EYA1 mutations leads to Branchio-Oto syndrome in two Chinese Han deaf families. Int J Pediatr Otorhinolaryngol. 2019; Aug. 123:141–5.35. Rickard S, Boxer M, Trompeter R, Bitner-Glindzicz M. Importance of clinical evaluation and molecular testing in the branchio-oto-renal (BOR) syndrome and overlapping phenotypes. J Med Genet. 2000; Aug. 37(8):623–7.36. Wang YG, Sun SP, Qiu YL, Xing QH, Lu W. A novel mutation in EYA1 in a Chinese family with Branchio-oto-renal syndrome. BMC Med Genet. 2018; Aug. 19(1):139.37. Henriksen AM, Tumer Z, Tommerup N, Tranebjaerg L, Larsen LA. Identification of a novel EYA1 splice-site mutation in a Danish branchio-oto-renal syndrome family. Genet Test. 2004; Winter. 8(4):404–6.38. Okada M, Fujimaru R, Morimoto N, Satomura K, Kaku Y, Tsuzuki K, et al. EYA1 and SIX1 gene mutations in Japanese patients with branchio-oto-renal (BOR) syndrome and related conditions. Pediatr Nephrol. 2006; Apr. 21(4):475–81.39. Sanggaard KM, Rendtorff ND, Kjaer KW, Eiberg H, Johnsen T, Gimsing S, et al. Branchio-oto-renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur J Hum Genet. 2007; Nov. 15(11):1121–31.40. Kim SH, Shin JH, Yeo CK, Chang SH, Park SY, Cho EH, et al. Identification of a novel mutation in the EYA1 gene in a Korean family with branchio-oto-renal (BOR) syndrome. Int J Pediatr Otorhinolaryngol. 2005; Aug. 69(8):1123–8.41. Gigante M, d'Altilia M, Montemurno E, Diella S, Bruno F, Netti GS, et al. Branchio-oto-renal syndrome (BOR) associated with focal glomerulosclerosis in a patient with a novel EYA1 splice site mutation. BMC Nephrol. 2013; Mar. 14:60.42. Castiglione A, Melchionda S, Carella M, Trevisi P, Bovo R, Manara R, et al. EYA1-related disorders: two clinical cases and a literature review. Int J Pediatr Otorhinolaryngol. 2014; Aug. 78(8):1201–10.43. Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, et al. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet. 1997; Dec. 6(13):2247–55.44. Kumar S, Kimberling WJ, Weston MD, Schaefer BG, Berg MA, Marres HA, et al. Identification of three novel mutations in human EYA1 protein associated with branchio-oto-renal syndrome. Hum Mutat. 1998; 11(6):443–9.45. Rodriguez-Soriano J, Vallo A, Bilbao JR, Castano L. Branchio-oto-renal syndrome: identification of a novel mutation in the EYA1 gene. Pediatr Nephrol. 2001; Jul. 16(7):550–3.46. Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019; Jan. 176(3):535–48.47. Ratnadiwakara M, Mohenska M, Anko ML. Splicing factors as regulators of miRNA biogenesis: links to human disease. Semin Cell Dev Biol. 2018; Jul. 79:113–22.48. Mango R, Biocca S, del Vecchio F, Clementi F, Sangiuolo F, Amati F, et al. In vivo and in vitro studies support that a new splicing isoform of OLR1 gene is protective against acute myocardial infarction. Circ Res. 2005; Jul. 97(2):152–8.49. Wang SH, Wu CC, Lu YC, Lin YH, Su YN, Hwu WL, et al. Mutation screening of the EYA1, SIX1, and SIX5 genes in an East Asian cohort with branchio-oto-renal syndrome. Laryngoscope. 2012; May. 122(5):1130–6.50. Lindau TA, Cardoso AC, Rossi NF, Giacheti CM. Anatomical changes and audiological profile in branchio-oto-renal syndrome: a literature review. Int Arch Otorhinolaryngol. 2014; Jan. 18(1):68–76.51. Kozlowski DJ, Whitfield TT, Hukriede NA, Lam WK, Weinberg ES. The zebrafish dog-eared mutation disrupts eya1, a gene required for cell survival and differentiation in the inner ear and lateral line. Dev Biol. 2005; Jan. 277(1):27–41.52. Christophorou NA, Bailey AP, Hanson S, Streit A. Activation of Six1 target genes is required for sensory placode formation. Dev Biol. 2009; Dec. 336(2):327–36.53. Soni UK, Roychoudhury K, Hegde RS. The Eyes Absent proteins in development and in developmental disorders. Biochem Soc Trans. 2021; Jun. 49(3):1397–408.54. Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997; Dec. 91(7):893–903.55. Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, et al. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999; Oct. 19(10):6815–24.56. Musharraf A, Kruspe D, Tomasch J, Besenbeck B, Englert C, Landgraf K. BOR-syndrome-associated Eya1 mutations lead to enhanced proteasomal degradation of Eya1 protein. PLoS One. 2014; Jan. 9(1):e87407.57. Kalatzis V, Sahly I, El-Amraoui A, Petit C. Eya1 expression in the developing ear and kidney: towards the understanding of the pathogenesis of Branchio-Oto-Renal (BOR) syndrome. Dev Dyn. 1998; Dec. 213(4):486–99.58. Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999; Sep. 23(1):113–7.59. Zheng Z, Wei X, Hildebrandt A, Schmidt B. A computational method for studying the relation between alternative splicing and DNA methylation. Nucleic Acids Res. 2016; Jan. 44(2):e19.60. Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011; Dec. 44(6):942–53.61. Hafiz A, Bakri R, Alsaad M, Fetni OM, Alsubaihi LI, Shamshad H. In silico prediction of Plasmodium falciparum cytoadherence inhibitors that disrupt interaction between gC1qR-DBLβ12 complex. Pharmaceuticals (Basel). 2022; May. 15(6):691.62. Srivastava I, Misra SK, Bangru S, Boateng KA, Soares JA, SchwartzDuval AS, et al. Complementary oligonucleotide conjugated multicolor carbon dots for intracellular recognition of biological events. ACS Appl Mater Interfaces. 2020; Apr. 12(14):16137–49.63. Ipsaro JJ, Huang L, Mondragon A. Structures of the spectrin-ankyrin interaction binding domains. Blood. 2009; May. 113(22):5385–93.64. Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nat Struct Mol Biol. 2013; Apr. 20(4):447–53.65. Goren A, Kim E, Amit M, Vaknin K, Kfir N, Ram O, et al. Overlapping splicing regulatory motifs: combinatorial effects on splicing. Nucleic Acids Res. 2010; Jun. 38(10):3318–27.66. Yang JY, Wang WQ, Han MY, Huang SS, Wang GJ, Su Y, et al. Addition of an affected family member to a previously ascertained autosomal recessive nonsyndromic hearing loss pedigree and systematic phenotype-genotype analysis of splice-site variants in MYO15A. BMC Med Genomics. 2022; Nov. 15(1):241.67. Gimsing S, Dyrmose J. Branchio-oto-renal dysplasia in three families. Ann Otol Rhinol Laryngol. 1986; Jul-Aug. 95(4 Pt 1):421–6.68. Cremers CW, Marres HA, Brunner HG. Neo-oval window technique and myringo-chorda-vestibulopexy in the BOR syndrome. Laryngoscope. 1993; Oct. 103(10):1186–9.69. Li HX, Zhou P, Tong M, Zheng Y. Branchial cleft fistula to branchiooto-renal syndrome: a case report and literature review. J Int Med Res. 2020; Jul. 48(7):300060520926363.70. Nam DW, Kang DW, Lee SM, Park MK, Lee JH, Oh SH, et al. Molecular genetic etiology and revisiting the middle ear surgery outcomes of branchio-oto-renal syndrome: experience in a tertiary referral center. Otol Neurotol. 2023; Jun. 44(5):e319–27.