Cancer Res Treat.
2023 Oct;55(4):1240-1249. 10.4143/crt.2022.1330.
Development and Validation of Models to Predict Lymph Node Metastasis in Early Gastric Cancer Using Logistic Regression and Gradient Boosting Machine Methods
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Pathology, Haeundae Paik Hospital, Inje University College of Medicine, Busan, Korea
- 3Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- 4Department of Surgery, Seoul National University Bundang Hospital, Seongnam, Korea
- KMID: 2547798
- DOI: http://doi.org/10.4143/crt.2022.1330
Abstract
- Purpose
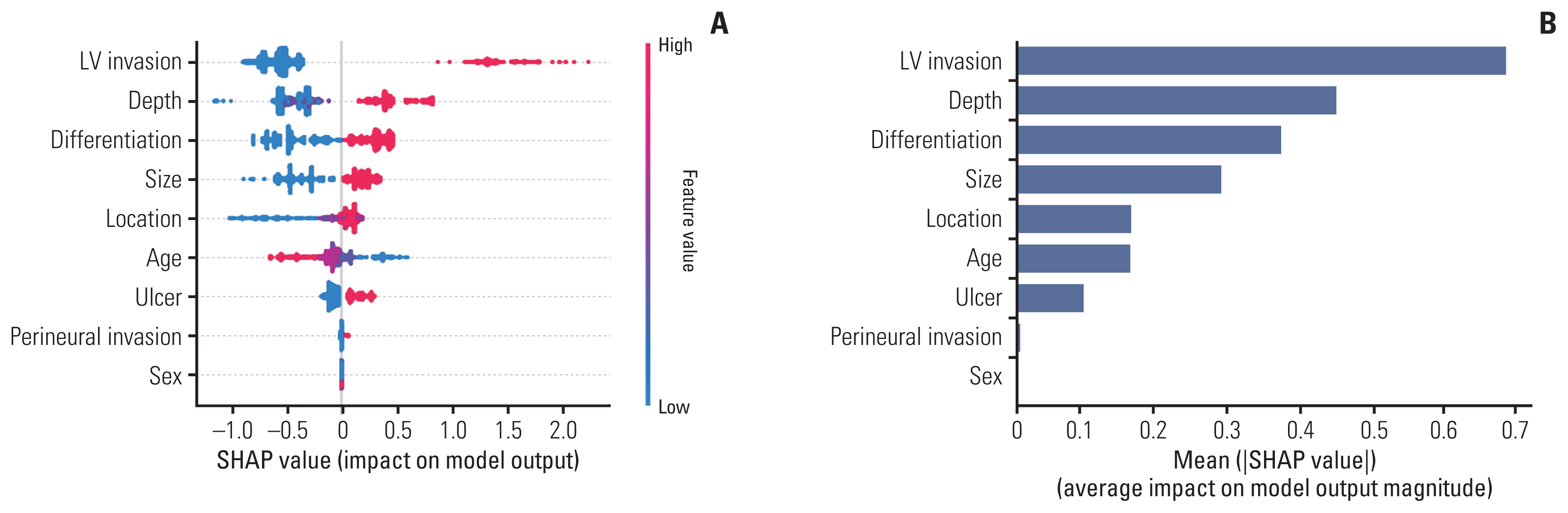
To identify important features of lymph node metastasis (LNM) and develop a prediction model for early gastric cancer (EGC) using a gradient boosting machine (GBM) method.
Materials and Methods
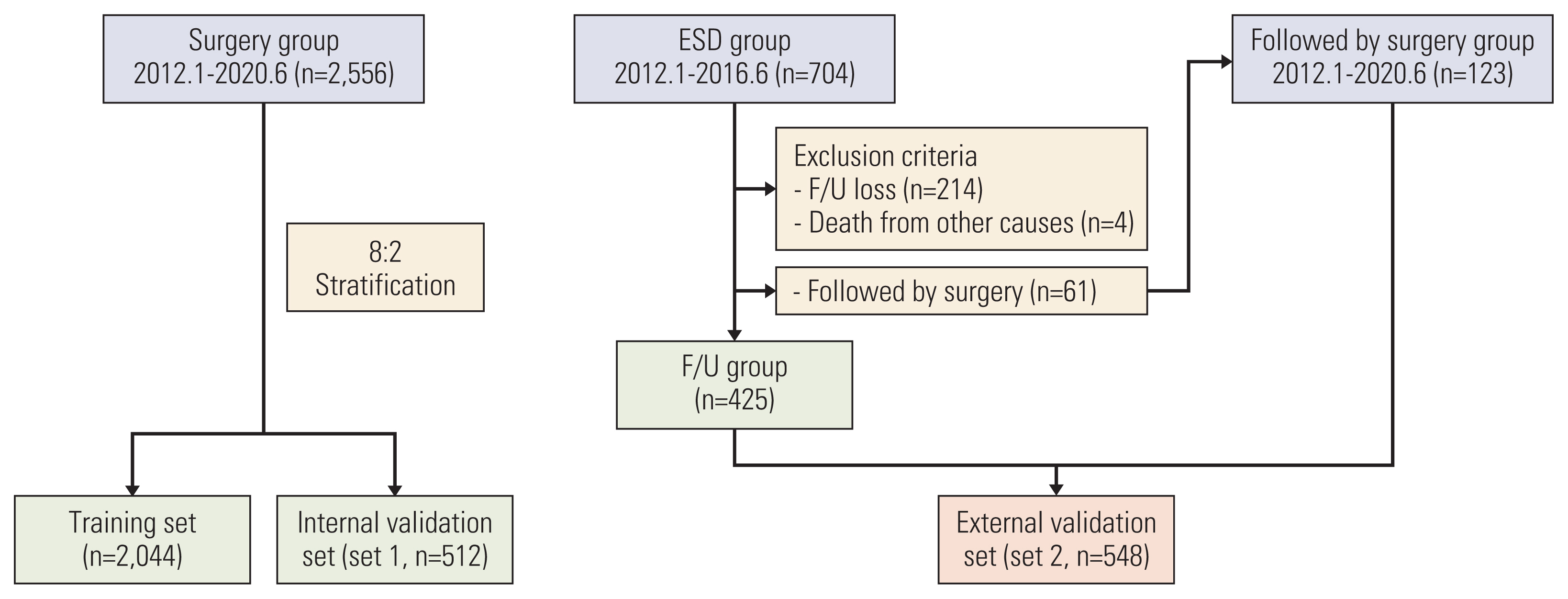
The clinicopathologic data of 2556 patients with EGC who underwent gastrectomy were used as training set and the internal validation set (set 1) at a ratio of 8:2. Additionally, 548 patients with EGC who underwent endoscopic submucosal dissection (ESD) as the initial treatment were included in the external validation set (set 2). The GBM model was constructed, and its performance was compared with that of the Japanese guidelines.
Results
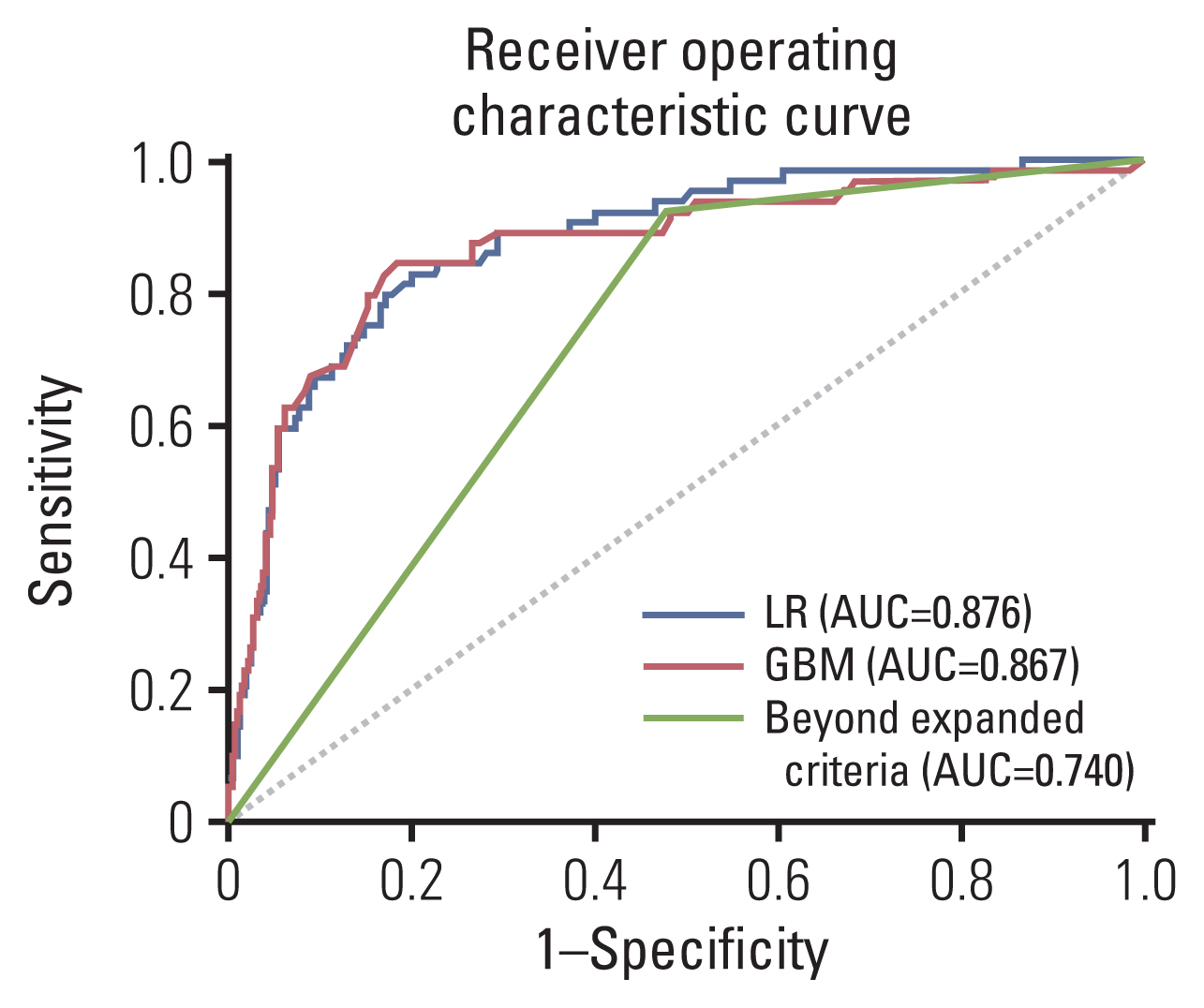
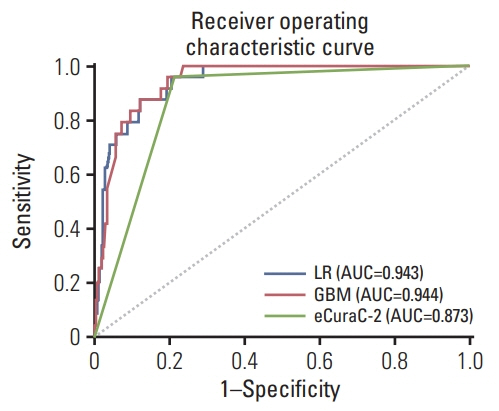
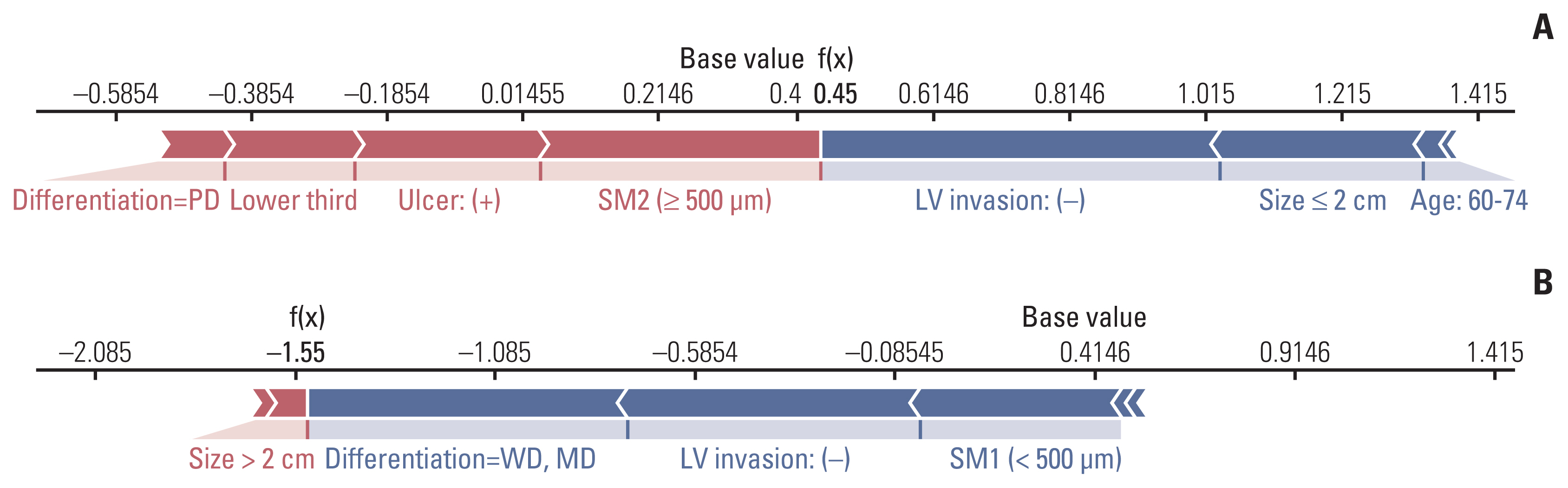
LNM was identified in 12.6% (321/2556) of the gastrectomy group (training set & set 1) and 4.3% (24/548) of the ESD group (set 2). In the GBM analysis, the top five features that most affected LNM were lymphovascular invasion, depth, differentiation, size, and location. The accuracy, sensitivity, specificity, and the area under the receiver operating characteristics of set 1 were 0.566, 0.922, 0.516, and 0.867, while those of set 2 were 0.810, 0.958, 0.803, and 0.944, respectively. When the sensitivity of GBM was adjusted to that of Japanese guidelines (beyond the expanded criteria in set 1 [0.922] and eCuraC-2 in set 2 [0.958]), the specificities of GBM in sets 1 and 2 were 0.516 (95% confidence interval, 0.502-0.523) and 0.803 (0.795-0.805), while those of the Japanese guidelines were 0.502 (0.488-0.509) and 0.788 (0.780-0.790), respectively.
Conclusion
The GBM model showed good performance comparable with the eCura system in predicting LNM risk in EGCs.
Figure
Reference
-
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.
Article2. Lee JH, Kim JG, Jung HK, Kim JH, Jeong WK, Jeon TJ, et al. Clinical practice guidelines for gastric cancer in Korea: an evidence-based approach. J Gastric Cancer. 2014; 14:87–104.
Article3. Ono H, Yao K, Fujishiro M, Oda I, Nimura S, Yahagi N, et al. Guidelines for endoscopic submucosal dissection and endoscopic mucosal resection for early gastric cancer. Dig Endosc. 2016; 28:3–15.
Article4. Ono H, Yao K, Fujishiro M, Oda I, Uedo N, Nimura S, et al. Guidelines for endoscopic submucosal dissection and endoscopic mucosal resection for early gastric cancer (second edition). Dig Endosc. 2021; 33:4–20.
Article5. Yamao T, Shirao K, Ono H, Kondo H, Saito D, Yamaguchi H, et al. Risk factors for lymph node metastasis from intramucosal gastric carcinoma. Cancer. 1996; 77:602–6.
Article6. Yasuda K, Shiraishi N, Suematsu T, Yamaguchi K, Adachi Y, Kitano S. Rate of detection of lymph node metastasis is correlated with the depth of submucosal invasion in early stage gastric carcinoma. Cancer. 1999; 85:2119–23.
Article7. Ren G, Cai R, Zhang WJ, Ou JM, Jin YN, Li WH. Prediction of risk factors for lymph node metastasis in early gastric cancer. World J Gastroenterol. 2013; 19:3096–107.
Article8. Kim SM, Lee H, Min BH, Kim JJ, An JY, Choi MG, et al. A prediction model for lymph node metastasis in early-stage gastric cancer: toward tailored lymphadenectomy. J Surg Oncol. 2019; 120:670–5.
Article9. Mu J, Jia Z, Yao W, Song J, Cao X, Jiang J, et al. Predicting lymph node metastasis in early gastric cancer patients: development and validation of a model. Future Oncol. 2019; 15:3609–17.
Article10. Pyo JH, Shin CM, Lee H, Min BH, Lee JH, Kim SM, et al. A Risk-prediction model based on lymph-node metastasis for incorporation into a treatment algorithm for signet ring cell-type intramucosal gastric cancer. Ann Surg. 2016; 264:1038–43.
Article11. Zheng Z, Zhang Y, Zhang L, Li Z, Wu X, Liu Y, et al. A nomogram for predicting the likelihood of lymph node metastasis in early gastric patients. BMC Cancer. 2016; 16:92.
Article12. Bang CS, Ahn JY, Kim JH, Kim YI, Choi IJ, Shin WG. Establishing machine learning models to predict curative resection in early gastric cancer with undifferentiated histology: development and usability study. J Med Internet Res. 2021; 23:e25053.
Article13. Kuo KM, Talley PC, Huang CH, Cheng LC. Predicting hospital-acquired pneumonia among schizophrenic patients: a machine learning approach. BMC Med Inform Decis Mak. 2019; 19:42.
Article14. Thio Q, Karhade AV, Ogink PT, Raskin KA, De Amorim Bernstein K, Lozano Calderon SA, et al. Can machine-learning techniques be used for 5-year survival prediction of patients with chondrosarcoma? Clin Orthop Relat Res. 2018; 476:2040–8.
Article15. Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019; 25:1666–83.
Article16. Zhou CM, Wang Y, Ye HT, Yan S, Ji M, Liu P, et al. Machine learning predicts lymph node metastasis of poorly differentiated-type intramucosal gastric cancer. Sci Rep. 2021; 11:1300.
Article17. Yun HR, Huh CW, Jung DH, Lee G, Son NH, Kim JH, et al. Machine learning improves the prediction rate of non-curative resection of endoscopic submucosal dissection in patients with early gastric cancer. Cancers (Basel). 2022; 14:3742.
Article18. Watson DS, Krutzinna J, Bruce IN, Griffiths CE, McInnes IB, Barnes MR, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ. 2019; 364:l886.
Article19. Dietterich TG. An experimental comparison of three methods for constructing ensembles of decision trees: bagging, boosting, and randomization. Mach Learn. 2000; 40:139–57.20. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12:2825–30.21. Chen T, Guestrin C. XGBoost. In : Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2016 Aug 13–17; San Francisco, CA, USA. New York: Association for Computing Machinery;2016.
Article22. Chu YN, Yu YN, Jing X, Mao T, Chen YQ, Zhou XB, et al. Feasibility of endoscopic treatment and predictors of lymph node metastasis in early gastric cancer. World J Gastroenterol. 2019; 25:5344–55.
Article23. Folli S, Morgagni P, Roviello F, De Manzoni G, Marrelli D, Saragoni L, et al. Risk factors for lymph node metastases and their prognostic significance in early gastric cancer (EGC) for the Italian Research Group for Gastric Cancer (IRGGC). Jpn J Clin Oncol. 2001; 31:495–9.
Article24. Lee JH, Choi IJ, Kook MC, Nam BH, Kim YW, Ryu KW. Risk factors for lymph node metastasis in patients with early gastric cancer and signet ring cell histology. Br J Surg. 2010; 97:732–6.
Article25. Zhu H, Wang G, Zheng J, Zhu H, Huang J, Luo E, et al. Preoperative prediction for lymph node metastasis in early gastric cancer by interpretable machine learning models: a multicenter study. Surgery. 2022; 171:1543–51.
Article26. Bibault JE, Chang DT, Xing L. Development and validation of a model to predict survival in colorectal cancer using a gradient-boosted machine. Gut. 2021; 70:884–9.
Article27. Kudo SE, Ichimasa K, Villard B, Mori Y, Misawa M, Saito S, et al. Artificial intelligence system to determine risk of T1 colorectal cancer metastasis to lymph node. Gastroenterology. 2021; 160:1075–84.
Article28. Arima N, Adachi K, Katsube T, Amano K, Ishihara S, Watanabe M, et al. Predictive factors for metachronous recurrence of early gastric cancer after endoscopic treatment. J Clin Gastroenterol. 1999; 29:44–7.
Article29. Na JE, Lee YC, Kim TJ, Lee H, Won HH, Min YW, et al. Machine learning model to stratify the risk of lymph node metastasis for early gastric cancer: a single-center cohort study. Cancers (Basel). 2022; 14:1121.
Article30. Zhang Z, Zhao Y, Canes A, Steinberg D, Lyashevska O; written on behalf of AME Big-Data Clinical Trial Collaborative Group. Predictive analytics with gradient boosting in clinical medicine. Ann Transl Med. 2019; 7:152.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A machine learning-based prediction model of pelvic lymph node metastasis in women with early-stage cervical cancer
- Significance of Lymph Node Metastasis in Early Gastric Cancer
- Development of Machine-Learning Models to Predict Ambulation Outcomes Following Spinal Metastasis Surgery
- Risk Factors Affecting Lymph Node Metastasis and Recurrence in Early Gastric Cancer
- Minimum Number of Retrieved Lymph Nodes for Staging in Gastric Cancer