Healthc Inform Res.
2023 Oct;29(4):286-300. 10.4258/hir.2023.29.4.286.
Named Entity Recognition in Electronic Health Records: A Methodological Review
- Affiliations
-
- 1Grupo de Investigación e Innovación Biomédica, Instituto Tecnológico Metropolitano, Antioquia, Colombia
- 2Facultad de Ingenierías, Universidad de Medellín, Antioquia, Colombia
- KMID: 2547682
- DOI: http://doi.org/10.4258/hir.2023.29.4.286
Abstract
Objectives
A substantial portion of the data contained in Electronic Health Records (EHR) is unstructured, often appearing as free text. This format restricts its potential utility in clinical decision-making. Named entity recognition (NER) methods address the challenge of extracting pertinent information from unstructured text. The aim of this study was to outline the current NER methods and trace their evolution from 2011 to 2022.
Methods
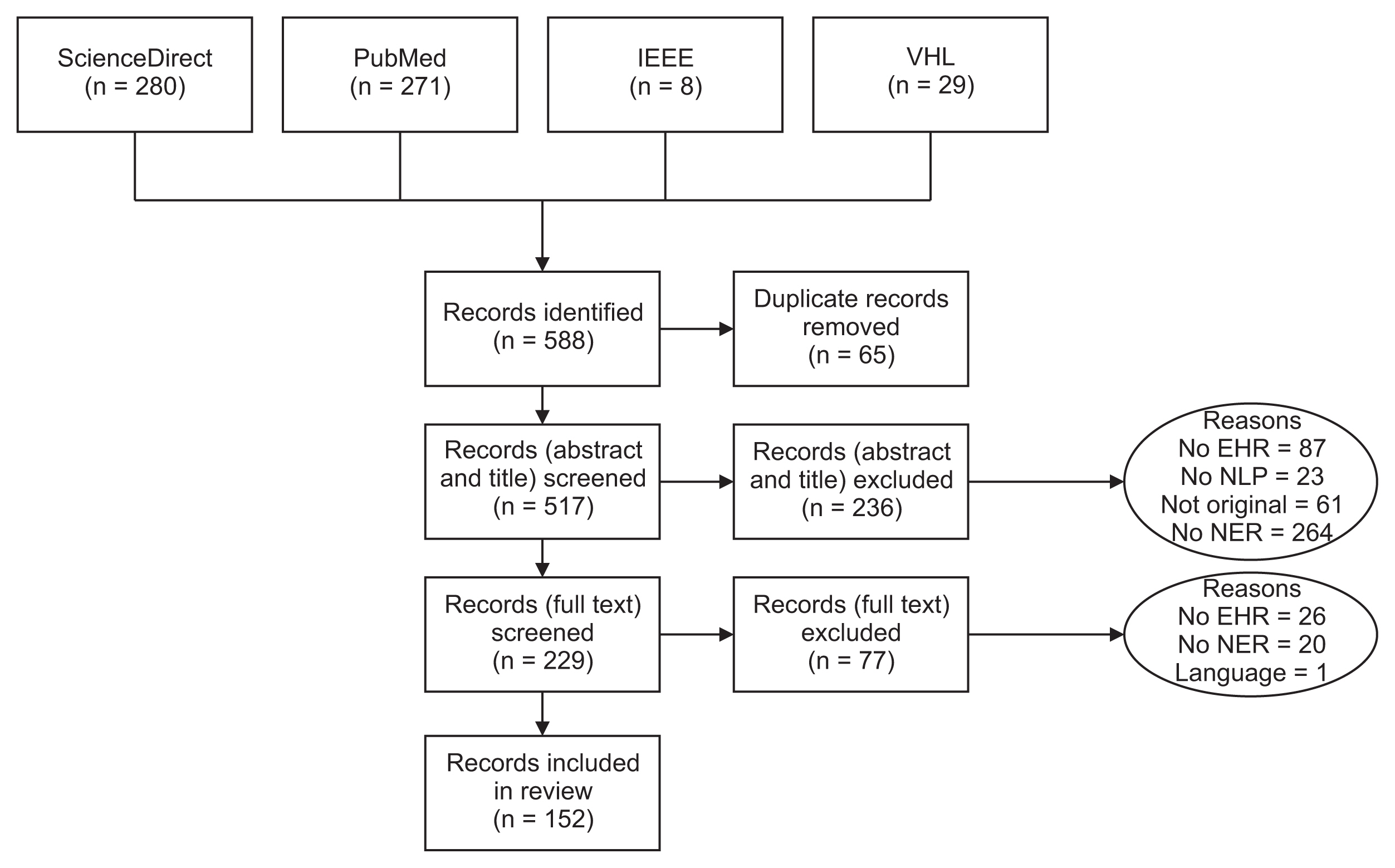
We conducted a methodological literature review of NER methods, with a focus on distinguishing the classification models, the types of tagging systems, and the languages employed in various corpora.
Results
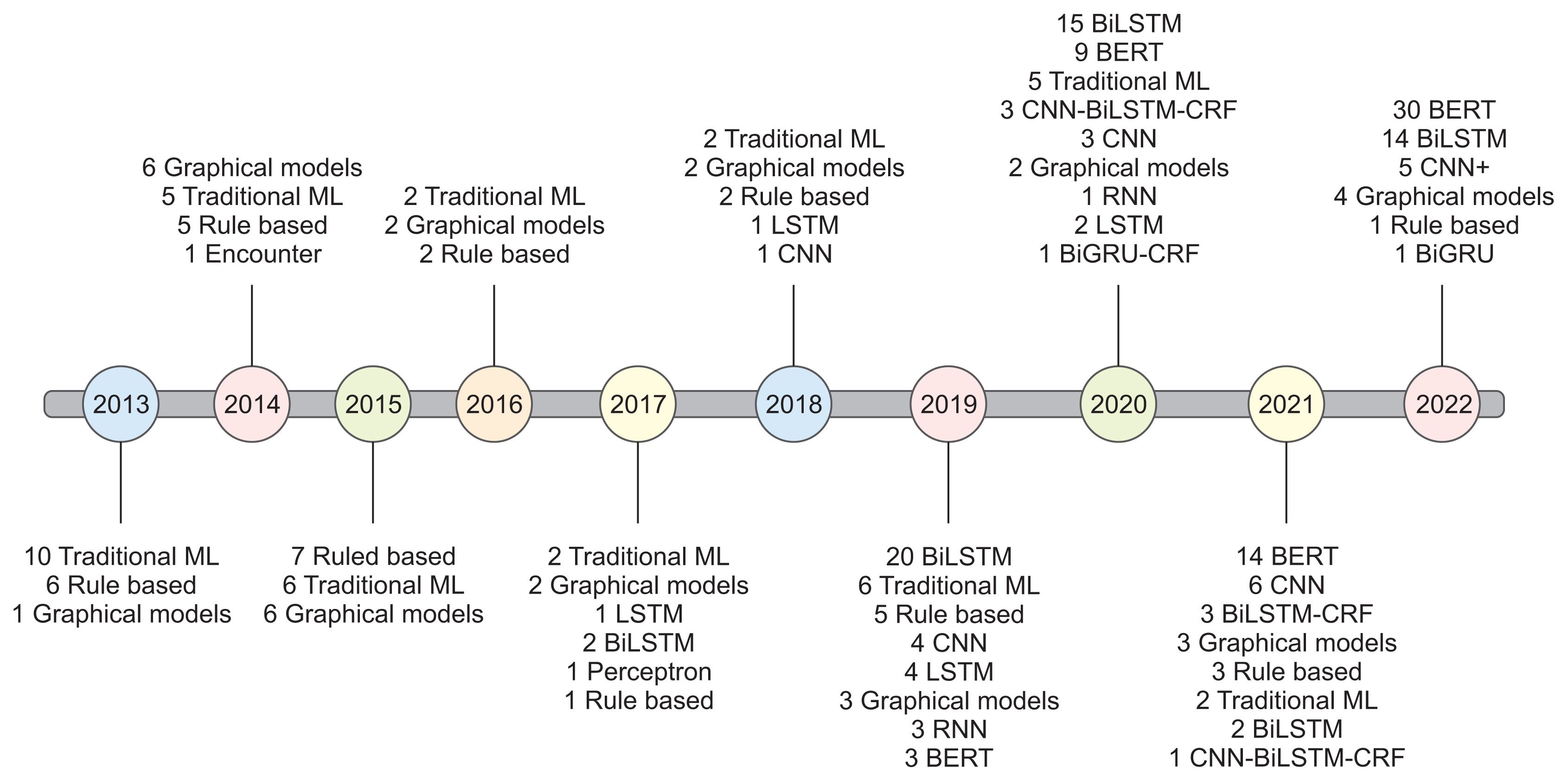
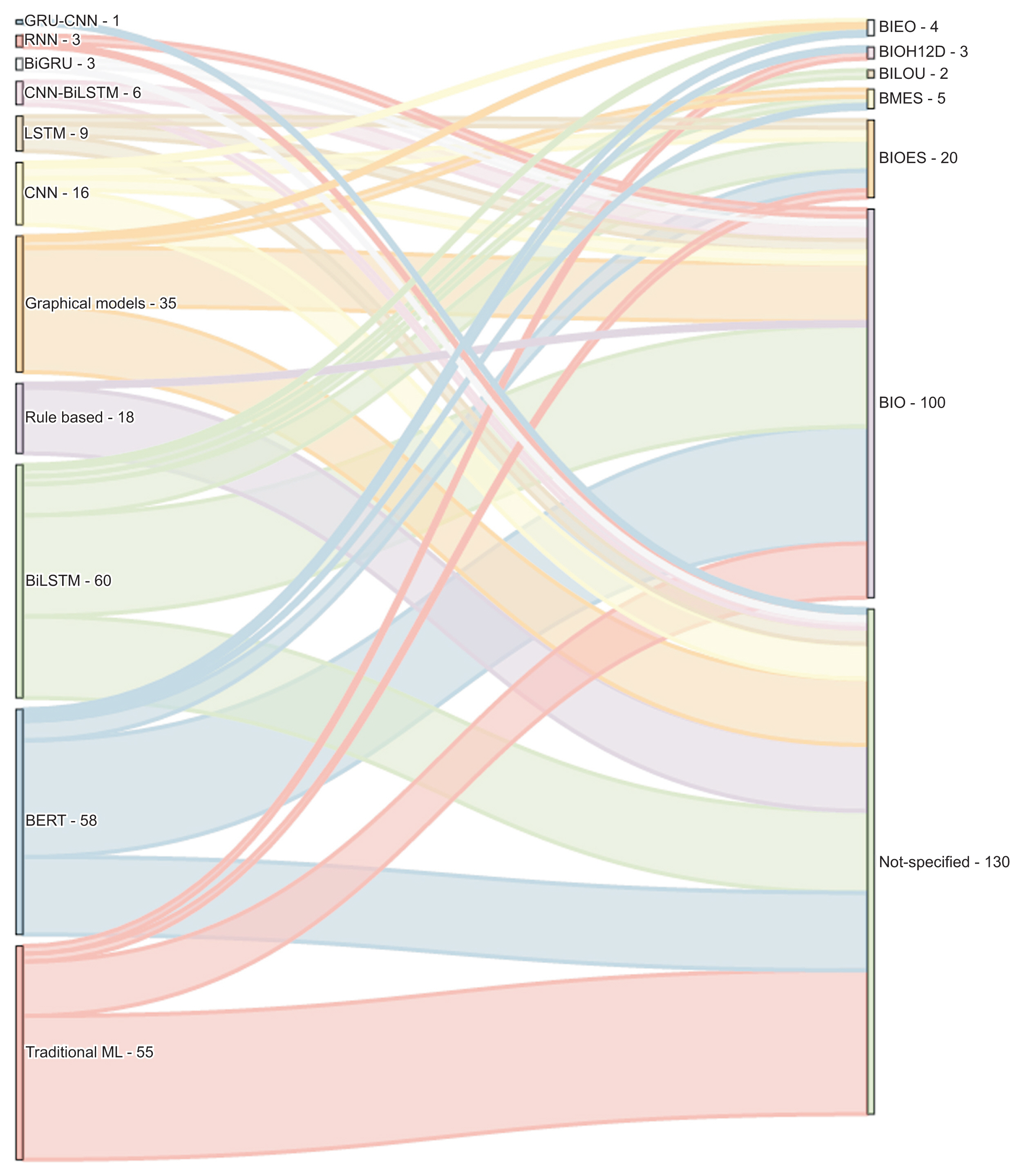
Several methods have been documented for automatically extracting relevant information from EHRs using natural language processing techniques such as NER and relation extraction (RE). These methods can automatically extract concepts, events, attributes, and other data, as well as the relationships between them. Most NER studies conducted thus far have utilized corpora in English or Chinese. Additionally, the bidirectional encoder representation from transformers using the BIO tagging system architecture is the most frequently reported classification scheme. We discovered a limited number of papers on the implementation of NER or RE tasks in EHRs within a specific clinical domain.
Conclusions
EHRs play a pivotal role in gathering clinical information and could serve as the primary source for automated clinical decision support systems. However, the creation of new corpora from EHRs in specific clinical domains is essential to facilitate the swift development of NER and RE models applied to EHRs for use in clinical practice.
Figure
Reference
-
References
1. Murdoch TB, Detsky AS.The inevitable application of big data to health care. JAMA. 2013; 309(13):1351–2. https://doi.org/10.1001/jama.2013.393.
Article2. Yehia E, Boshnak H, AbdelGaber S, Abdo A, Elzanfaly DS.Ontology-based clinical information extraction from physician’s free-text notes. J Biomed Inform. 2019; 98:103276. https://doi.org/10.1016/j.jbi.2019.103276.
Article3. ElDin HG, AbdulRazek M, Abdelshafi M, Sahlol AT.Med-Flair: medical named entity recognition for diseases and medications based on Flair embedding. Procedia Comput Sci. 2021; 189:67–75. https://doi.org/10.1016/j.procs.2021.05.078.
Article4. Kaplar A, Stosovic M, Kaplar A, Brkovic V, Naumovic R, Kovacevic A.Evaluation of clinical named entity recognition methods for Serbian electronic health records. Int J Med Inform. 2022; 164:104805. https://doi.org/10.1016/j.ijmedinf.2022.104805.
Article5. Neuraz A, Looten V, Rance B, Daniel N, Garcelon N, Llanos LC, et al. Do you need embeddings trained on a massive specialized corpus for your clinical natural language processing task? Stud Health Technol Inform. 2019; 264:1558–9. https://doi.org/10.3233/shti190533.
Article6. Kong J, Zhang L, Jiang M, Liu T.Incorporating multi-level CNN and attention mechanism for Chinese clinical named entity recognition. J Biomed Inform. 2021; 116:103737. https://doi.org/10.1016/j.jbi.2021.103737.
Article7. Xiong Y, Wang Z, Jiang D, Wang X, Chen Q, Xu H, et al. A fine-grained Chinese word segmentation and part-of-speech tagging corpus for clinical text. BMC Med Inform Decis Mak. 2019; 19(Suppl 2):66. https://doi.org/10.1186/s12911-019-0770-7.
Article8. Jindal P, Roth D.Extraction of events and temporal expressions from clinical narratives. J Biomed Inform. 2013; 46 (Suppl):S13–9. https://doi.org/10.1016/j.jbi.2013.08.010.
Article9. Jiang M, Chen Y, Liu M, Rosenbloom ST, Mani S, Denny JC, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc. 2011; 18(5):601–6. https://doi.org/10.1136/amiajnl-2011-000163.
Article10. Nikfarjam A, Emadzadeh E, Gonzalez G. Towards generating a patient’s timeline: extracting temporal relationships from clinical notes. J Biomed Inform. 2013. 46(Suppl 0):S40–7. https://doi.org/10.1016/j.jbi.2013.11.001.
Article11. Casillas A, Perez A, Oronoz M, Gojenola K, Santiso S.Learning to extract adverse drug reaction events from electronic health records in Spanish. Expert Syst Appl. 2016; 61(1):235–45. https://doi.org/10.1016/j.eswa.2016.05.034.
Article12. Henriksson A, Kvist M, Dalianis H, Duneld M.Identifying adverse drug event information in clinical notes with distributional semantic representations of context. J Biomed Inform. 2015; 57:333–49. https://doi.org/10.1016/j.jbi.2015.08.013.
Article13. de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X.Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc. 2011; 18(5):557–62. https://doi.org/10.1136/amiajnl-2011-000150.
Article14. Cormack J, Nath C, Milward D, Raja K, Jonnalagadda SR. Agile text mining for the 2014 i2b2/UTHealth Cardiac risk factors challenge. J Biomed Inform. 2015. 58:Suppl0. S120–7. https://doi.org/10.1016/j.jbi.2015.06.030.
Article15. Grouin C, Grabar N, Hamon T, Rosset S, Tannier X, Zweigenbaum P.Eventual situations for timeline extraction from clinical reports. J Am Med Inform Assoc. 2013; 20(5):820–7. https://doi.org/10.1136/amiajnl-2013-001627.
Article16. Lei J, Tang B, Lu X, Gao K, Jiang M, Xu H.A comprehensive study of named entity recognition in Chinese clinical text. J Am Med Inform Assoc. 2014; 21(5):808–14. https://doi.org/10.1136/amiajnl-2013-002381.
Article17. Dligach D, Bethard S, Becker L, Miller T, Savova GK.Discovering body site and severity modifiers in clinical texts. J Am Med Inform Assoc. 2014; 21(3):448–54. https://doi.org/10.1136/amiajnl-2013-001766.
Article18. Jung K, LePendu P, Iyer S, Bauer-Mehren A, Percha B, Shah NH.Functional evaluation of out-of-the-box text-mining tools for data-mining tasks. J Am Med Inform Assoc. 2015; 22(1):121–31. https://doi.org/10.1136/amiajnl-2014-002902.
Article19. Yang H, Garibaldi JM.Automatic detection of protected health information from clinic narratives. J Biomed Inform. 2015; 58(Suppl):S30–8. https://doi.org/10.1016%2Fj.jbi.2015.06.015.
Article20. Lee W, Kim K, Lee EY, Choi J.Conditional random fields for clinical named entity recognition: a comparative study using Korean clinical texts. Comput Biol Med. 2018; 101:7–14. https://doi.org/10.1016/j.compbiomed.2018.07.019.
Article21. Wang H, Zhang W, Zeng Q, Li Z, Feng K, Liu L.Extracting important information from Chinese Operation Notes with natural language processing methods. J Biomed Inform. 2014; 48:130–6. https://doi.org/10.1016/j.jbi.2013.12.017.
Article22. Grouin C, Neveol A.De-identification of clinical notes in French: towards a protocol for reference corpus development. J Biomed Inform. 2014; 50:151–61. https://doi.org/10.1016/j.jbi.2013.12.014.
Article23. Kovacevic A, Dehghan A, Filannino M, Keane JA, Nenadic G.Combining rules and machine learning for extraction of temporal expressions and events from clinical narratives. J Am Med Inform Assoc. 2013; 20(5):859–66. https://doi.org/10.1136/amiajnl-2013-001625.
Article24. Xiong Y, Peng H, Xiang Y, Wong KC, Chen Q, Yan J, et al. Leveraging Multi-source knowledge for Chinese clinical named entity recognition via relational graph convolutional network. J Biomed Inform. 2022; 128:104035. https://doi.org/10.1016/j.jbi.2022.104035.
Article25. Jiale N, Gao D, Sun Y, Li X, Shen X, Li M, et al. Surgical procedure long terms recognition from Chinese literature incorporating structural feature. Heliyon. 2022; 8(11):e11291. https://doi.org/10.1016/j.heliyon.2022.e11291.
Article26. Hassanpour S, Langlotz CP.Information extraction from multi-institutional radiology reports. Artif Intell Med. 2016; 66:29–39. https://doi.org/10.1016/j.artmed.2015.09.007.
Article27. Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging [Internet]. Ithaca (NY): arXiv. org;2015. [cited at 2023 Sep 30]. Available from: https://arxiv.org/abs/1508.01991.28. Zhang X, Zhang Y, Zhang Q, Ren Y, Qiu T, Ma J, et al. Extracting comprehensive clinical information for breast cancer using deep learning methods. Int J Med Inform. 2019; 132:103985. https://doi.org/10.1016/j.ijmedinf.2019.103985.
Article29. Xu J, Li Z, Wei Q, Wu Y, Xiang Y, Lee HJ, et al. Applying a deep learning-based sequence labeling approach to detect attributes of medical concepts in clinical text. BMC Med Inform Decis Mak. 2019; 19(Suppl 5):236. https://doi.org/10.1186/s12911-019-0937-2.
Article30. Li PL, Yuan ZM, Tu WN, Yu K, Lu DX.Medical knowledge extraction and analysis from electronic medical records using deep learning. Chin Med Sci J. 2019; 34(2):133–9. https://doi.org/10.24920/003589.
Article31. Suarez-Paniagua V, Rivera Zavala RM, Segura-Bedmar I, Martinez P.A two-stage deep learning approach for extracting entities and relationships from medical texts. J Biomed Inform. 2019; 99:103285. https://doi.org/10.1016/j.jbi.2019.103285.
Article32. Weegar R, Perez A, Casillas A, Oronoz M.Recent advances in Swedish and Spanish medical entity recognition in clinical texts using deep neural approaches. BMC Med Inform Decis Mak. 2019; 19(Suppl 7):274. https://doi.org/10.1186/s12911-019-0981-y.
Article33. Ji B, Liu R, Li S, Yu J, Wu Q, Tan Y, et al. A hybrid approach for named entity recognition in Chinese electronic medical record. BMC Med Inform Decis Mak. 2019; 19(Suppl 2):64. https://doi.org/10.1186/s12911-019-0767-2.
Article34. Wang Q, Zhou Y, Ruan T, Gao D, Xia Y, He P.Incorporating dictionaries into deep neural networks for the Chinese clinical named entity recognition. J Biomed Inform. 2019; 92:103133. https://doi.org/10.1016/j.jbi.2019.103133.
Article35. Yin M, Mou C, Xiong K, Ren J.Chinese clinical named entity recognition with radical-level feature and selfattention mechanism. J Biomed Inform. 2019; 98:103289. https://doi.org/10.1016/j.jbi.2019.103289.
Article36. Alex B, Grover C, Tobin R, Sudlow C, Mair G, Whiteley W.Text mining brain imaging reports. J Biomed Semantics. 2019; 10(Suppl 1):23. http://dx.doi.org/10.1186/s13326-019-0211-7.
Article37. Shi X, Yi Y, Xiong Y, Tang B, Chen Q, Wang X, et al. Extracting entities with attributes in clinical text via joint deep learning. J Am Med Inform Assoc. 2019; 26(12):1584–91. https://doi.org/10.1093/jamia/ocz158.
Article38. Wang Y, Ananiadou S, Tsujii J.Improving clinical named entity recognition in Chinese using the graphical and phonetic feature. BMC Med Inform Decis Mak. 2019; 19(Suppl 7):273. https://doi.org/10.1186/s12911-019-0980-z.
Article39. Casillas A, Ezeiza N, Goenaga I, Perez A, Soto X.Measuring the effect of different types of unsupervised word representations on Medical Named Entity Recognition. Int J Med Inform. 2019; 129:100–6. https://doi.org/10.1016/j.ijmedinf.2019.05.022.
Article40. Chen Y, Zhou C, Li T, Wu H, Zhao X, Ye K, et al. Named entity recognition from Chinese adverse drug event reports with lexical feature based BiLSTM-CRF and tri-training. J Biomed Inform. 2019; 96:103252. https://doi.org/10.1016/j.jbi.2019.103252.
Article41. Liu X, Zhou Y, Wang Z.Recognition and extraction of named entities in online medical diagnosis data based on a deep neural network. J Vis Commun Image Represent. 2019; 60:1–15. https://doi.org/10.1016/j.jvcir.2019.02.001.
Article42. Li Z, Yang J, Gou X, Qi X.Recurrent neural networks with segment attention and entity description for relation extraction from clinical texts. Artif Intell Med. 2019; 97:9–18. https://doi.org/10.1016/j.artmed.2019.04.003.
Article43. Juckett DA, Kasten EP, Davis FN, Gostine M.Concept detection using text exemplars aligned with a specialized ontology. Data Knowl Eng. 2019; 119:22–35. https://doi.org/10.1016/j.datak.2018.11.002.
Article44. Su J, Hu J, Jiang J, Xie J, Yang Y, He B, et al. Extraction of risk factors for cardiovascular diseases from Chinese electronic medical records. Comput Methods Programs Biomed. 2019; 172:1–10. https://doi.org/10.1016/j.cmpb.2019.01.007.
Article45. Li L, Zhao J, Hou L, Zhai Y, Shi J, Cui F.An attention-based deep learning model for clinical named entity recognition of Chinese electronic medical records. BMC Med Inform Decis Mak. 2019; 19(Suppl 5):235. https://doi.org/10.1186/s12911-019-0933-6.
Article46. Dong X, Chowdhury S, Qian L, Li X, Guan Y, Yang J, et al. Deep learning for named entity recognition on Chinese electronic medical records: Combining deep transfer learning with multitask bi-directional LSTM RNN. PLoS One. 2019; 14(5):e0216046. https://doi.org/10.1371/journal.pone.0216046.
Article47. Lin CH, Hsu KC, Liang CK, Lee TH, Liou CW, Lee JD, et al. A disease-specific language representation model for cerebrovascular disease research. Comput Methods Programs Biomed. 2021; 211:106446. https://doi.org/10.1016/j.cmpb.2021.106446.
Article48. Murugadoss K, Rajasekharan A, Malin B, Agarwal V, Bade S, Anderson JR, et al. Building a best-in-class automated de-identification tool for electronic health records through ensemble learning. Patterns (N Y). 2021; 2(6):100255. https://doi.org/10.1016/j.patter.2021.100255.
Article49. Harnoune A, Rhanoui M, Mikram M, Yousfi S, Elkaimbillah Z, El Asri B.BERT based clinical knowledge extraction for biomedical knowledge graph construction and analysis. Comput Methods Programs Biomed Update. 2021; 1:100042. https://doi.org/10.1016/j.cmpbup.2021.100042.
Article50. Narayanan S, Achan P, Rangan PV, Rajan SP.Unified concept and assertion detection using contextual multitask learning in a clinical decision support system. J Biomed Inform. 2021; 122:103898. https://doi.org/10.1016/j.jbi.2021.103898.
Article51. Thieu T, Maldonado JC, Ho PS, Ding M, Marr A, Brandt D, et al. A comprehensive study of mobility functioning information in clinical notes: entity hierarchy, corpus annotation, and sequence labeling. Int J Med Inform. 2021; 147:104351. https://doi.org/10.1016/j.ijmedinf.2020.104351liu.
Article52. Liu X, Liu Y, Wu H, Guan Q.A tag based joint extraction model for Chinese medical text. Comput Biol Chem. 2021; 93:107508. https://doi.org/10.1016/j.compbiolchem.2021.107508.
Article53. Puccetti G, Chiarello F, Fantoni G.A simple and fast method for Named Entity context extraction from patents. Expert Syst Appl. 2021; 184:115570. https://doi.org/10.1016/j.eswa.2021.115570.
Article54. El-allaly ED, Sarrouti M, En-Nahnahi N, El Alaoui SO.MTTLADE: a multi-task transfer learning-based method for adverse drug events extraction. Inf Process Manag. 2021; 58(3):102473. https://doi.org/10.1016/j.ipm.2020.102473.
Article55. Uronen L, Salantera S, Hakala K, Hartiala J, Moen H.Combining supervised and unsupervised named entity recognition to detect psychosocial risk factors in occupational health checks. Int J Med Inform. 2022; 160:104695. https://doi.org/10.1016/j.ijmedinf.2022.104695.
Article56. Xiong Y, Peng W, Chen Q, Huang Z, Tang B.A unified machine reading comprehension framework for cohort selection. IEEE J Biomed Health Inform. 2022; 26(1):379–87. https://doi.org/10.1109/jbhi.2021.3095478.
Article57. Zhang B, Liu K, Wang H, Li M, Pan J.Chinese named-entity recognition via self-attention mechanism and position-aware influence propagation embedding. Data Knowl Eng. 2022; 139:101983. https://doi.org/10.1016/j.datak.2022.101983.
Article58. Landolsi MY, Ben Romdhane L, Hlaoua L. Medical named entity recognition using surrounding sequences matching. Procedia Comput Sci. 2022. 207:674–83. https://doi.org/10.1016/j.procs.2022.09.122.
Article59. Shi J, Sun M, Sun Z, Li M, Gu Y, Zhang W.Multi-level semantic fusion network for Chinese medical named entity recognition. J Biomed Inform. 2022; 133:104144. https://doi.org/10.1016/j.jbi.2022.104144.
Article60. Gerardin C, Wajsburt P, Vaillant P, Bellamine A, Carrat F, Tannier X.Multilabel classification of medical concepts for patient clinical profile identification. Artif Intell Med. 2022; 128:102311. https://doi.org/10.1016/j.artmed.2022.102311.
Article61. Madan S, Julius Zimmer F, Balabin H, Schaaf S, Frohlich H, Fluck J, et al. Deep learning-based detection of psychiatric attributes from German mental health records. Int J Med Inform. 2022; 161:104724. https://doi.org/10.1016/j.ijmedinf.2022.104724.
Article62. Narayanan S, Madhuri SS, Ramesh MV, Rangan PV, Rajan SP.Deep contextual multi-task feature fusion for enhanced concept, negation and speculation detection from clinical notes. Informatics Med Unlocked. 2022; 34:101109. https://doi.org/10.1016/j.imu.2022.101109.
Article63. An Y, Xia X, Chen X, Wu FX, Wang J.Chinese clinical named entity recognition via multi-head self-attention based BiLSTM-CRF. Artif Intell Med. 2022; 127:102282. https://doi.org/10.1016/j.artmed.2022.102282.
Article64. Wang SY, Huang J, Hwang H, Hu W, Tao S, Hernandez-Boussard T.Leveraging weak supervision to perform named entity recognition in electronic health records progress notes to identify the ophthalmology exam. Int J Med Inform. 2022; 167:104864. https://doi.org/10.1016/j.ijmedinf.2022.104864.
Article65. Narayanan S, Mannam K, Achan P, Ramesh MV, Rangan PV, Rajan SP.A contextual multi-task neural approach to medication and adverse events identification from clinical text. J Biomed Inform. 2022; 125:103960. https://doi.org/10.1016/j.jbi.2021.103960.
Article66. El-Allaly ED, Sarrouti M, En-Nahnahi N, Ouatik El Alaoui S.An attentive joint model with transformer-based weighted graph convolutional network for extracting adverse drug event relation. J Biomed Inform. 2022; 125:103968. https://doi.org/10.1016/j.jbi.2021.103968.
Article67. Sun J, Liu Y, Cui J, He H.Deep learning-based methods for natural hazard named entity recognition. Sci Rep. 2022; 12(1):4598. https://doi.org/10.1038/s41598-022-08667-2.
Article68. Fang A, Hu J, Zhao W, Feng M, Fu J, Feng S, et al. Extracting clinical named entity for pituitary adenomas from Chinese electronic medical records. BMC Med Inform Decis Mak. 2022; 22(1):72. https://doi.org/10.1186/s12911-022-01810-z.
Article69. Zhou S, Wang N, Wang L, Liu H, Zhang R.Cancer-BERT: a cancer domain-specific language model for extracting breast cancer phenotypes from electronic health records. J Am Med Inform Assoc. 2022; 29(7):1208–16. https://doi.org/10.1093/jamia/ocac040.
Article70. Guo S, Yang W, Han L, Song X, Wang G.A multi-layer soft lattice based model for Chinese clinical named entity recognition. BMC Med Inform Decis Mak. 2022; 22(1):201. https://doi.org/10.1186/s12911-022-01924-4.
Article71. Lee EB, Heo GE, Choi CM, Song M.MLM-based typographical error correction of unstructured medical texts for named entity recognition. BMC Bioinformatics. 2022; 23(1):486. https://doi.org/10.1186/s12859-022-05035-9.
Article72. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017; 30:5999–6009.73. Xu Y, Wang Y, Liu T, Liu J, Fan Y, Qian Y, et al. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. J Am Med Inform Assoc. 2014; 21(e1):e84–92. https://doi.org/10.1136/amiajnl-2013-001806.
Article74. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020; 36(4):1234–40. https://doi.org/10.48550/arXiv.1901.08746.
Article75. Peng Y, Yan S, Lu Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets [Internet]. Ithaca (NY): arXiv.org;2019. [cited at 2023 Sep 30]. Available from: https://arxiv.org/abs/1906.05474.76. Ji B, Li S, Yu J, Ma J, Tang J, Wu Q, et al. Research on Chinese medical named entity recognition based on collaborative cooperation of multiple neural network models. J Biomed Inform. 2020; 104:103395.
Article77. Sterckx L, Vandewiele G, Dehaene I, Janssens O, Ongenae F, De Backere F, et al. Clinical information extraction for preterm birth risk prediction. J Biomed Inform. 2020; 110:103544. https://doi.org/10.1016/j.jbi.2020.103395.
Article78. Kormilitzin A, Vaci N, Liu Q, Nevado-Holgado A.Med7: a transferable clinical natural language processing model for electronic health records. Artif Intell Med. 2021; 118:102086. https://doi.org/10.1016/j.artmed.2021.102086.
Article79. Wei Q, Ji Z, Li Z, Du J, Wang J, Xu J, et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc. 2020; 27(1):13–21. https://doi.org/10.1093/jamia/ocz063.
Article80. Khan S, Shamsi JA.Health Quest: a generalized clinical decision support system with multi-label classification. J King Saud Univ–Comput Inf Sci. 2021; 33(1):45–53. https://doi.org/10.1016/j.jksuci.2018.11.003.
Article81. Lin WC, Chen JS, Kaluzny J, Chen A, Chiang MF, Hribar MR.Extraction of active medications and adherence using natural language processing for glaucoma patients. AMIA Annu Symp Proc. 2021; 2021:773–82.82. Dai HJ.Family member information extraction via neural sequence labeling models with different tag schemes. BMC Med Inform Decis Mak. 2019; 19(Suppl 10):257.
Article83. Chen L, Li Y, Chen W, Liu X, Yu Z, Zhang S.Utilizing soft constraints to enhance medical relation extraction from the history of present illness in electronic medical records. J Biomed Inform. 2018; 87:108–17.
Article84. Dai HJ, Su CH, Wu CS.Adverse drug event and medication extraction in electronic health records via a cascading architecture with different sequence labeling models and word embeddings. J Am Med Inform Assoc. 2020; 27(1):47–55. https://doi.org/10.1093/jamia/ocz120.
Article85. Yang X, Bian J, Fang R, Bjarnadottir RI, Hogan WR, Wu Y.Identifying relations of medications with adverse drug events using recurrent convolutional neural networks and gradient boosting. J Am Med Inform Assoc. 2020; 27(1):65–72. https://doi.org/10.1093/jamia/ocz144.
Article86. Li L, Xu W, Yu H.Character-level neural network model based on Nadam optimization and its application in clinical concept extraction. Neurocomputing. 2020; 414:182–90. https://doi.org/10.1016/j.neucom.2020.07.027.
Article87. Zhang R, Zhao P, Guo W, Wang R, Lu W.Medical named entity recognition based on dilated convolutional neural network. Cogn Robot. 2022; 2:13–20. https://doi.org/10.1016/j.cogr.2021.11.002.
Article88. Cheng Y, Anick P, Hong P, Xue N.Temporal relation discovery between events and temporal expressions identified in clinical narrative. J Biomed Inform. 2013; 46 (Suppl):S48–53. https://doi.org/10.1016/j.jbi.2013.09.010.
Article89. Liu Z, Yang M, Wang X, Chen Q, Tang B, Wang Z, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak. 2017; 17(Suppl 2):67. https://doi.org/10.1186%2Fs12911-017-0468-7.
Article90. Kochmar E, Andersen O, Briscoe T. HOO 2012 error recognition and correction shared task: Cambridge University submission report. In : Proceedings of the 7th Workshop on Innovative Use of NLP for Building Educational Applications; 2012 Jun 7; Montreal, Canada. p. 242–50. https://doi.org/10.17863/CAM.9671.
Article91. Zhao S, Cai Z, Chen H, Wang Y, Liu F, Liu A.Adversarial training based lattice LSTM for Chinese clinical named entity recognition. J Biomed Inform. 2019; 99:103290. https://doi.org/10.1016/j.jbi.2019.103290.
Article92. Yu G, Yang Y, Wang X, Zhen H, He G, Li Z, et al. Adversarial active learning for the identification of medical concepts and annotation inconsistency. J Biomed Inform. 2020; 108:103481. https://doi.org/10.1016/j.jbi.2020.103481.
Article93. Wang C, Wang H, Zhuang H, Li W, Han S, Zhang H, et al. Chinese medical named entity recognition based on multi-granularity semantic dictionary and multimodal tree. J Biomed Inform. 2020; 111:103583. https://doi.org/10.1016/j.jbi.2020.103583.
Article94. Chen X, Ouyang C, Liu Y, Bu Y.Improving the named entity recognition of Chinese electronic medical records by combining domain dictionary and rules. Int J Environ Res Public Health. 2020; 17(8):2687. https://doi.org/10.3390/ijerph17082687.
Article95. Kersloot MG, Lau F, Abu-Hanna A, Arts DL, Cornet R.Automated SNOMED CT concept and attribute relationship detection through a web-based implementation of cTAKES. J Biomed Semantics. 2019; 10(1):14. https://doi.org/10.1186/s13326-019-0207-3.
Article96. Christopoulou F, Tran TT, Sahu SK, Miwa M, Ananiadou S.Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. J Am Med Inform Assoc. 2020; 27(1):39–46.
Article97. Hu Y, Ameer I, Zuo W, Peng X, Zhou Y, Li Z, et al. Zeroshot clinical entity recognition using ChatGPT [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2023 Sep 30]. Available from: https://arxiv.org/abs/2303.16416.98. Li X, Zhu X, Ma Z, Liu X, Shas S. Are ChatGPT and GPT-4 general-purpose solvers for financial text analytics? An examination on several typical tasks [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2023 Sep 30]. Available from: https://arxiv.org/abs/2305.05862.99. Lai VD, Ngo NT, Veyseh AP, Man H, Dernoncourt F, Bui T, et al. ChatGPT beyond English: towards a comprehensive evaluation of large language models in multilingual learning [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2023 Sep 30]. Available from: https://arxiv.org/abs/2304.05613.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Improving classification of low-resource COVID-19 literature by using Named Entity Recognition
- OryzaGP 2021 update: a rice gene and protein dataset for named-entity recognition
- Improving spaCy dependency annotation and PoS tagging web service using independent NER services
- Using the PubAnnotation ecosystem to perform agile text mining on Genomics & Informatics: a tutorial review
- Clinical Terminologies: A Solution for Semantic Interoperability