Korean J Physiol Pharmacol.
2023 Sep;27(5):449-456. 10.4196/kjpp.2023.27.5.449.
N-retinylidene-N-retinylethanolamine degradation in human retinal pigment epithelial cells via memantine- and ifenprodil-mediated autophagy
- Affiliations
-
- 1College of Pharmacy, Gachon Research Institute of Pharmaceutical Sciences, Gachon University, Incheon 21936, Korea
- KMID: 2545535
- DOI: http://doi.org/10.4196/kjpp.2023.27.5.449
Abstract
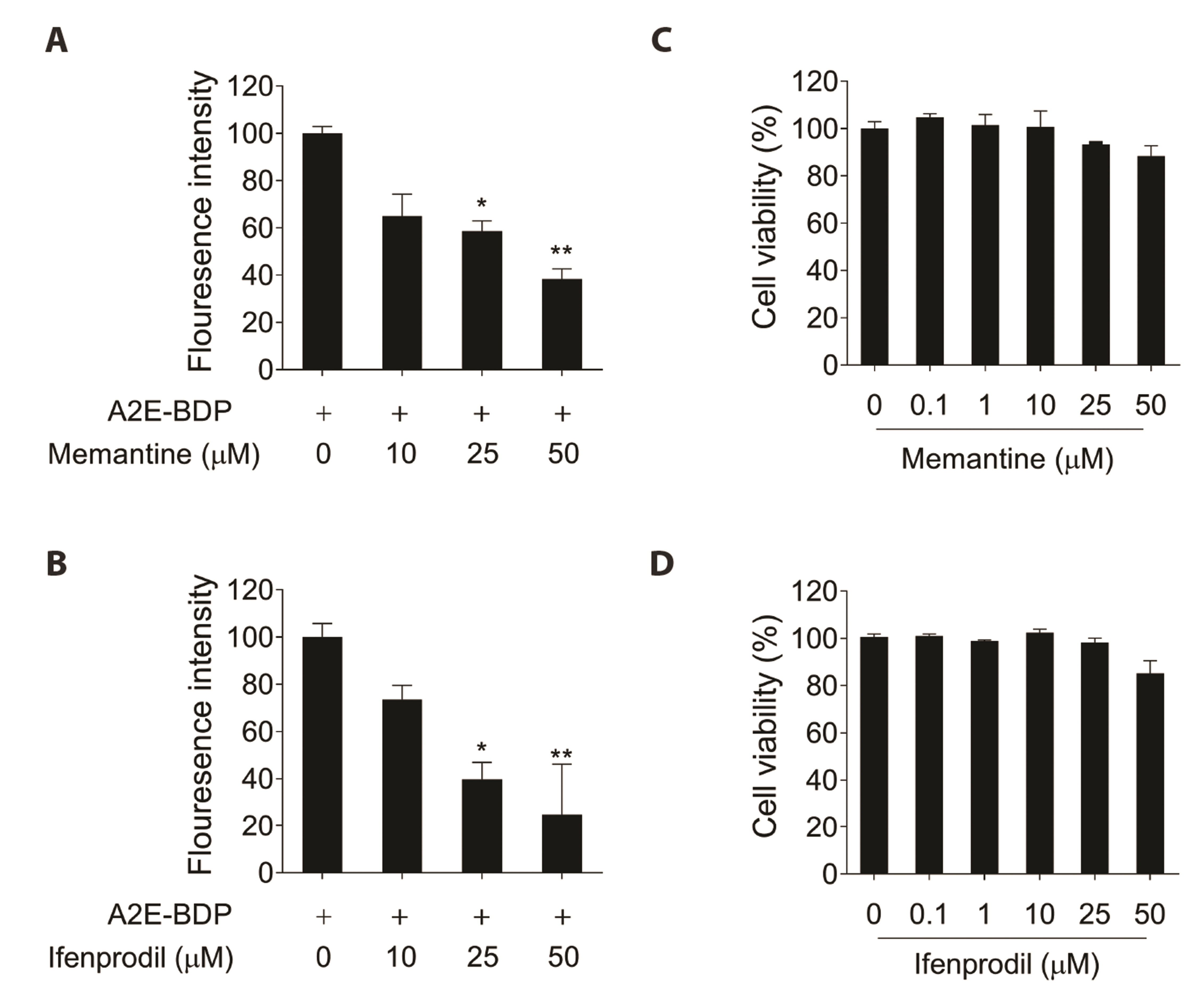
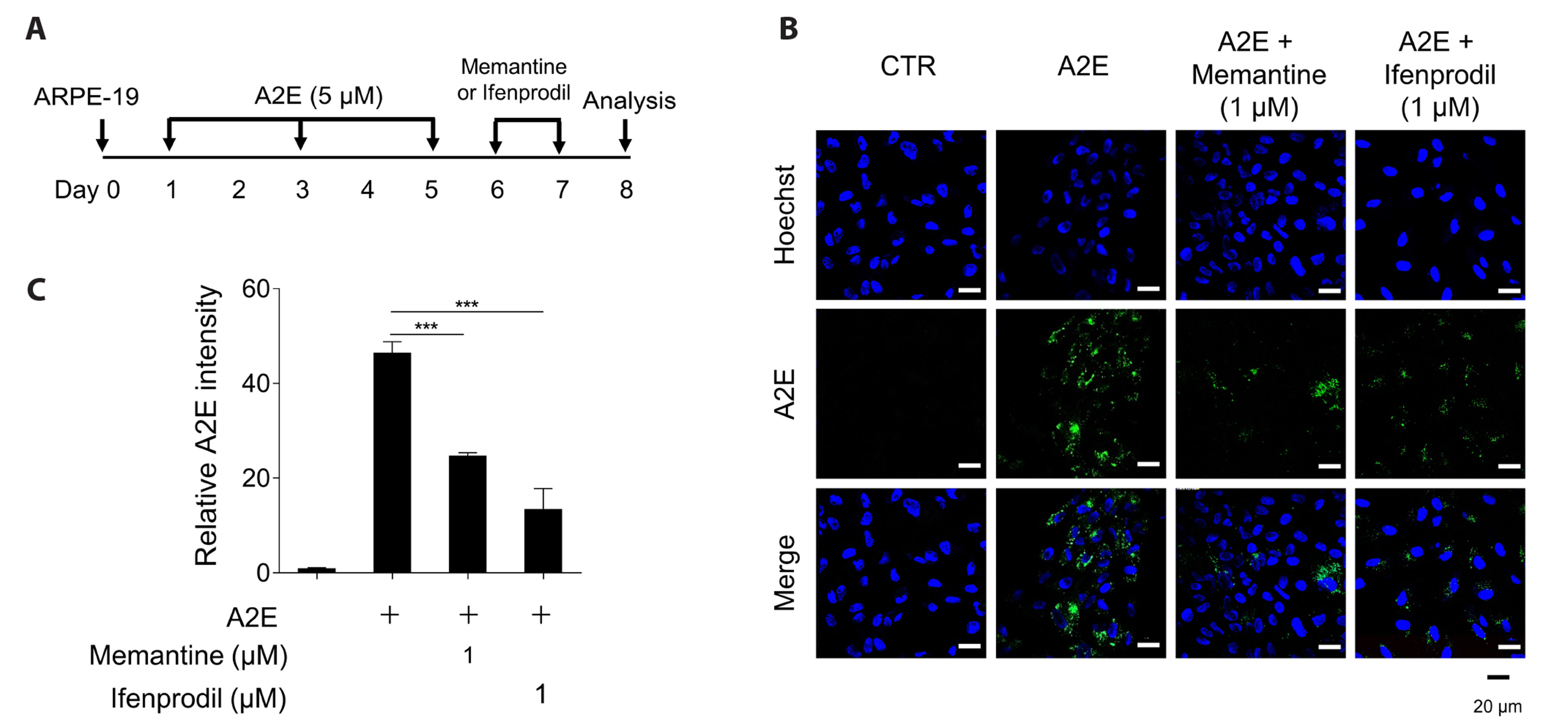
- N-methyl-D-aspartate (NMDA) receptors are ionic glutamine receptors involved in brain development and functions such as learning and memory formation. NMDA receptor inhibition is associated with autophagy activation. In this study, we investigated whether the NMDA receptor antagonists, memantine and ifenprodil, induce autophagy in human retinal pigment epithelial cells (ARPE-19) to remove Nretinylidene-N-retinylethanolamine (A2E), an intracellular lipofuscin component. Fluorometric analysis using labeled A2E (A2E-BDP) and confocal microscopic examination revealed that low concentrations of NMDA receptor antagonists, which did not induce cytotoxicity, significantly reduced A2E accumulation in ARPE-19 cells. In addition, memantine and ifenprodil activated autophagy in ARPE-19 cells as measured by microtubule-associated protein 1A/1B-light chain3-II formation and phosphorylated p62 protein levels. Further, to understand the correlation between memantine- and ifenprodil-mediated A2E degradation and autophagy, autophagy-related 5 (ATG5) was depleted using RNA interference. Memantine and ifenprodil failed to degrade A2E in ARPE-19 cells lacking ATG5. Taken together, our study indicates that the NMDA receptor antagonists, memantine and ifenprodil, can remove A2E accumulated in cells via autophagy activation in ARPE-19 cells.
Keyword
Figure
Reference
-
1. Laube B, Hirai H, Sturgess M, Betz H, Kuhse J. 1997; Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron. 18:493–503. DOI: 10.1016/S0896-6273(00)81249-0. PMID: 9115742.
Article2. Yau SY, Bettio L, Vetrici M, Truesdell A, Chiu C, Chiu J, Truesdell E, Christie BR. 2018; Chronic minocycline treatment improves hippocampal neuronal structure, NMDA receptor function, and memory processing in Fmr1 knockout mice. Neurobiol Dis. 113:11–22. DOI: 10.1016/j.nbd.2018.01.014. PMID: 29367010.
Article3. Korinek M, Kapras V, Vyklicky V, Adamusova E, Borovska J, Vales K, Stuchlik A, Horak M, Chodounska H, Vyklicky L Jr. 2011; Neurosteroid modulation of N-methyl-D-aspartate receptors: molecular mechanism and behavioral effects. Steroids. 76:1409–1418. DOI: 10.1016/j.steroids.2011.09.002. PMID: 21925193.
Article4. Świetlik D, Kusiak A, Ossowska A. 2022; Computational modeling of therapy with the NMDA antagonist in neurodegenerative disease: information theory in the mechanism of action of memantine. Int J Environ Res Public Health. 19:4727. DOI: 10.3390/ijerph19084727. PMID: 35457595. PMCID: PMC9027074.
Article5. Lipton SA. 2007; Pathologically-activated therapeutics for neuroprotection: mechanism of NMDA receptor block by memantine and S-nitrosylation. Curr Drug Targets. 8:621–632. DOI: 10.2174/138945007780618472. PMID: 17504105.
Article6. Johnson JW, Kotermanski SE. 2006; Mechanism of action of memantine. Curr Opin Pharmacol. 6:61–67. DOI: 10.1016/j.coph.2005.09.007. PMID: 16368266.
Article7. Hirano K, Fujimaki M, Sasazawa Y, Yamaguchi A, Ishikawa KI, Miyamoto K, Souma S, Furuya N, Imamichi Y, Yamada D, Saya H, Akamatsu W, Saiki S, Hattori N. 2019; Neuroprotective effects of memantine via enhancement of autophagy. Biochem Biophys Res Commun. 518:161–170. DOI: 10.1016/j.bbrc.2019.08.025. PMID: 31431260.
Article8. Companys-Alemany J, Turcu AL, Schneider M, Müller CE, Vázquez S, Griñán-Ferré C, Pallàs M. 2022; NMDA receptor antagonists reduce amyloid-β deposition by modulating calpain-1 signaling and autophagy, rescuing cognitive impairment in 5XFAD mice. Cell Mol Life Sci. 79:408. DOI: 10.1007/s00018-022-04438-4. PMID: 35810220. PMCID: PMC9271115.
Article9. Trivedi PC, Bartlett JJ, Pulinilkunnil T. 2020; Lysosomal biology and function: modern view of cellular debris bin. Cells. 9:1131. DOI: 10.3390/cells9051131. PMID: 32375321. PMCID: PMC7290337. PMID: c1e053a9d2a14302ab5abf8ecd22d51c.
Article10. Mizushima N, Komatsu M. 2011; Autophagy: renovation of cells and tissues. Cell. 147:728–741. DOI: 10.1016/j.cell.2011.10.026. PMID: 22078875.
Article11. Bechthold E, Schreiber JA, Lehmkuhl K, Frehland B, Schepmann D, Bernal FA, Daniliuc C, Álvarez I, Garcia CV, Schmidt TJ, Seebohm G, Wünsch B. 2021; Ifenprodil stereoisomers: synthesis, absolute configuration, and correlation with biological activity. J Med Chem. 64:1170–1179. DOI: 10.1021/acs.jmedchem.0c01912. PMID: 33426889.
Article12. Sun JY, Zhao SJ, Wang HB, Hou YJ, Mi QJ, Yang MF, Yuan H, Ni QB, Sun BL, Zhang ZY. 2021; Ifenprodil improves long-term neurologic deficits through antagonizing glutamate-induced excitotoxicity after experimental subarachnoid hemorrhage. Transl Stroke Res. 12:1067–1080. DOI: 10.1007/s12975-021-00906-4. PMID: 33713028.
Article13. Sparrow JR, Cai B, Fishkin N, Jang YP, Krane S, Vollmer HR, Zhou J, Nakanishi K. 2003; A2E, a fluorophore of RPE lipofuscin: can it cause RPE degeneration? Adv Exp Med Biol. 533:205–211. DOI: 10.1007/978-1-4615-0067-4_26. PMID: 15180266.
Article14. Gray DA, Woulfe J. 2005; Lipofuscin and aging: a matter of toxic waste. Sci Aging Knowledge Environ. 2005:re1. DOI: 10.1126/sageke.2005.1.nf1. PMID: 15689603.
Article15. Li WW, Wang HJ, Tan YZ, Wang YL, Yu SN, Li ZH. 2021; Reducing lipofuscin accumulation and cardiomyocytic senescence of aging heart by enhancing autophagy. Exp Cell Res. 403:112585. DOI: 10.1016/j.yexcr.2021.112585. PMID: 33811905.
Article16. Sparrow JR, Boulton M. 2005; RPE lipofuscin and its role in retinal pathobiology. Exp Eye Res. 80:595–606. DOI: 10.1016/j.exer.2005.01.007. PMID: 15862166.
Article17. Shin CY, Lee MH, Kim HM, Chung HC, Kim DU, Lee JH, Jeong KW. 2022; Protective effect of Ribes nigrum extract against blue light-induced retinal degeneration in vitro and in vivo. Antioxidants (Basel). 11:832. DOI: 10.3390/antiox11050832. PMID: 35624696. PMCID: PMC9137918. PMID: e82a44894c0b49ebadba1b15b6fa51d0.
Article18. Pham TNM, Shin CY, Park SH, Lee TH, Ryu HY, Kim SB, Auh K, Jeong KW. 2021; Solanum melongena L. extract protects retinal pigment epithelial cells from blue light-induced phototoxicity in in vitro and in vivo models. Nutrients. 13:359. DOI: 10.3390/nu13020359. PMID: 33503991. PMCID: PMC7912168. PMID: 8e3cdbaf95044e9e8d461c594ec07736.
Article19. Jin HL, Lee SC, Kwon YS, Choung SY, Jeong KW. 2016; A novel fluorescence-based assay for measuring A2E removal from human retinal pigment epithelial cells to screen for age-related macular degeneration inhibitors. J Pharm Biomed Anal. 117:560–567. DOI: 10.1016/j.jpba.2015.10.010. PMID: 26604166.
Article20. Lee JR, Jeong KW. 2022; NMDA receptor antagonists degrade lipofuscin via autophagy in human retinal pigment epithelial cells. Medicina (Kaunas). 58:1129. DOI: 10.3390/medicina58081129. PMID: 36013596. PMCID: PMC9415004. PMID: 491fe915b916435bb2c5fe79b4547973.
Article21. Mizushima N, Yoshimori T, Levine B. 2010; Methods in mammalian autophagy research. Cell. 140:313–326. DOI: 10.1016/j.cell.2010.01.028. PMID: 20144757. PMCID: PMC2852113.
Article22. Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. 2000; A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492. DOI: 10.1038/35044114. PMID: 11100732.
Article23. Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. 2013; Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 51:618–631. DOI: 10.1016/j.molcel.2013.08.003. PMID: 24011591.
Article24. Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. 2012; Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 109:13561–13566. DOI: 10.1073/pnas.1121572109. PMID: 22872865. PMCID: PMC3427110.
Article25. Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, Komatsu M, Oh YJ, Zhao Y, Yue Z. 2015; Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 11:e1004987. DOI: 10.1371/journal.pgen.1004987. PMID: 25723488. PMCID: PMC4344198. PMID: 5a1d09fcc7f04c35a1f002ff307bde9a.
Article26. Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. 2011; Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 44:279–289. DOI: 10.1016/j.molcel.2011.07.039. PMID: 22017874.
Article27. Ye X, Zhou XJ, Zhang H. 2018; Exploring the role of autophagy-related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front Immunol. 9:2334. DOI: 10.3389/fimmu.2018.02334. PMID: 30386331. PMCID: PMC6199349. PMID: fdff729608b54260b70e8c9b12c1b310.28. Ishibashi K, Fujita N, Kanno E, Omori H, Yoshimori T, Itoh T, Fukuda M. 2011; Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy. 7:1500–1513. DOI: 10.4161/auto.7.12.18025. PMID: 22082872. PMCID: PMC3288023.
Article29. Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. 2008; The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 19:2092–2100. DOI: 10.1091/mbc.e07-12-1257. PMID: 18321988. PMCID: PMC2366860.
Article30. Ikonomidou C, Turski L. 2002; Why did NMDA receptor antagonists fail clinical trials for stroke and traumatic brain injury? Lancet Neurol. 1:383–386. DOI: 10.1016/S1474-4422(02)00164-3. PMID: 12849400.
Article31. Winkler D, Leyhe T. 2018; Alzheimer's disease - State of the art, and emerging diagnostics and therapeutics. Ther Umsch. 75:432–437. German. DOI: 10.1024/0040-5930/a001020. PMID: 30935362.32. Klein R, Klein BE, Knudtson MD, Meuer SM, Swift M, Gangnon RE. 2007; Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 114:253–262. DOI: 10.1016/j.ophtha.2006.10.040. PMID: 17270675.
Article33. Baird PN, Robman LD, Richardson AJ, Dimitrov PN, Tikellis G, McCarty CA, Guymer RH. 2008; Gene-environment interaction in progression of AMD: the CFH gene, smoking and exposure to chronic infection. Hum Mol Genet. 17:1299–1305. DOI: 10.1093/hmg/ddn018. PMID: 18203751.
Article34. Hammer M, Richter S, Guehrs KH, Schweitzer D. 2006; Retinal pigment epithelium cell damage by A2-E and its photo-derivatives. Mol Vis. 12:1348–1354. DOI: 10.1007/springerreference_36603. PMID: 17110917.35. Shin CY, Jeong KW. 2022; Photooxidation of A2E by blue light regulates heme oxygenase 1 expression via NF-κB and lysine methyltransferase 2A in ARPE-19 cells. Life (Basel). 12:1698. DOI: 10.3390/life12111698. PMID: 36362853. PMCID: PMC9699413. PMID: a991e08b9c50406085871c58ffe72a08.
Article36. Jeong SY, Gu X, Jeong KW. 2019; Photoactivation of N-retinylidene-N-retinylethanolamine compromises autophagy in retinal pigmented epithelial cells. Food Chem Toxicol. 131:110555. DOI: 10.1016/j.fct.2019.06.002. PMID: 31173818.
Article37. Jin HL, Jeong KW. 2022; Transcriptome analysis of long-term exposure to blue light in retinal pigment epithelial cells. Biomol Ther (Seoul). 30:291–297. DOI: 10.4062/biomolther.2021.155. PMID: 35074938. PMCID: PMC9047491.
Article38. Yoon WS, Yeom MY, Kang ES, Chung YA, Chung DS, Jeun SS. 2017; Memantine induces NMDAR1-mediated autophagic cell death in malignant glioma cells. J Korean Neurosurg Soc. 60:130–137. DOI: 10.3340/jkns.2016.0101.006. PMID: 28264232. PMCID: PMC5365296.
Article39. Yao Y, Ju P, Liu H, Wu X, Niu Z, Zhu Y, Zhang C, Fang Y. 2020; Ifenprodil rapidly ameliorates depressive-like behaviors, activates mTOR signaling and modulates proinflammatory cytokines in the hippocampus of CUMS rats. Psychopharmacology (Berl). 237:1421–1433. DOI: 10.1007/s00213-020-05469-0. PMID: 32130432.
Article40. Al-Bari MAA, Xu P. 2020; Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann N Y Acad Sci. 1467:3–20. DOI: 10.1111/nyas.14305. PMID: 31985829.
Article41. Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. 2009; Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16:46–56. DOI: 10.1038/cdd.2008.110. PMID: 18636076.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Growth Patterns of Human Retinal Pigment Epithelium in Vitro
- Transcriptome Analysis of Long-Term Exposure to Blue Light in Retinal Pigment Epithelial Cells
- The Effect of 5-Fluorouraci1 on the Activity of the Retinal Pigment Epithelium in Vitro
- Culture of Retinal Pigment Epithelial Cells on Collagen Membrane
- Comparison of Inhibitory Effects of 5-Fluorouracil and Mitomycin C on Proliferation of Fibroblagts and Retinal Pigment Epithelial Cells