Cancer Res Treat.
2023 Jan;55(1):155-166. 10.4143/crt.2021.1567.
Genomic Signatures from Clinical Tumor Sequencing in Patients with Breast Cancer Having Germline BRCA1/2 Mutation
- Affiliations
-
- 1Division of Medical Oncology and Hematology, Department of Internal Medicine, Korea University Anam Hospital, Seoul, Korea
- 2K-MASTER Cancer Precision Medicine Diagnosis and Treatment Enterprise, Korea University Medical Center, Seoul, Korea
- 3Division of Endocrinology, Department of Internal Medicine, Korea University Anam Hospital, Seoul, Korea
- 4Department of Laboratory Medicine, Korea University Anam Hospital, Seoul, Korea
- 5Department of Breast Surgery, Korea University Anam Hospital, Seoul, Korea
- KMID: 2538001
- DOI: http://doi.org/10.4143/crt.2021.1567
Abstract
- Purpose
BRCA1 and BRCA2 are among the most important genes involved in DNA repair via homologous recombination (HR). Germline BRCA1/2 (gBRCA1/2)-related cancers have specific characteristics and treatment options but conducting gBRCA1/2 testing and interpreting the genetic imprint are sometimes complicated. Here, we describe the concordance of gBRCA1/2 derived from a panel of clinical tumor tissues using next-generation sequencing (NGS) and genetic aspects of tumors harboring gBRCA1/2 pathogenic variants.
Materials and Methods
Targeted sequencing was performed using available tumor tissue from patients who underwent gBRCA1/2 testing. Comparative genomic analysis was performed according to gBRCA1/2 pathogenicity.
Results
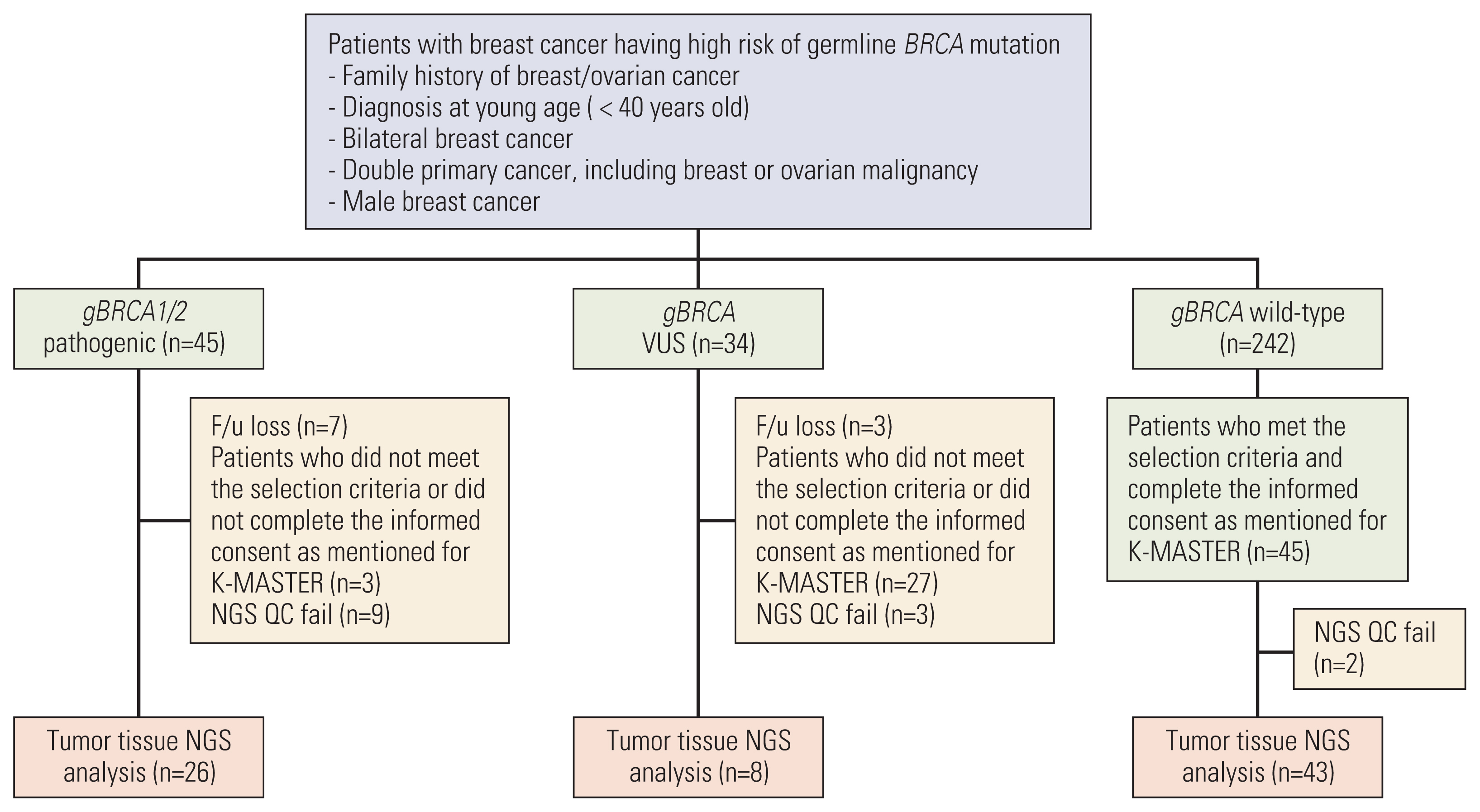
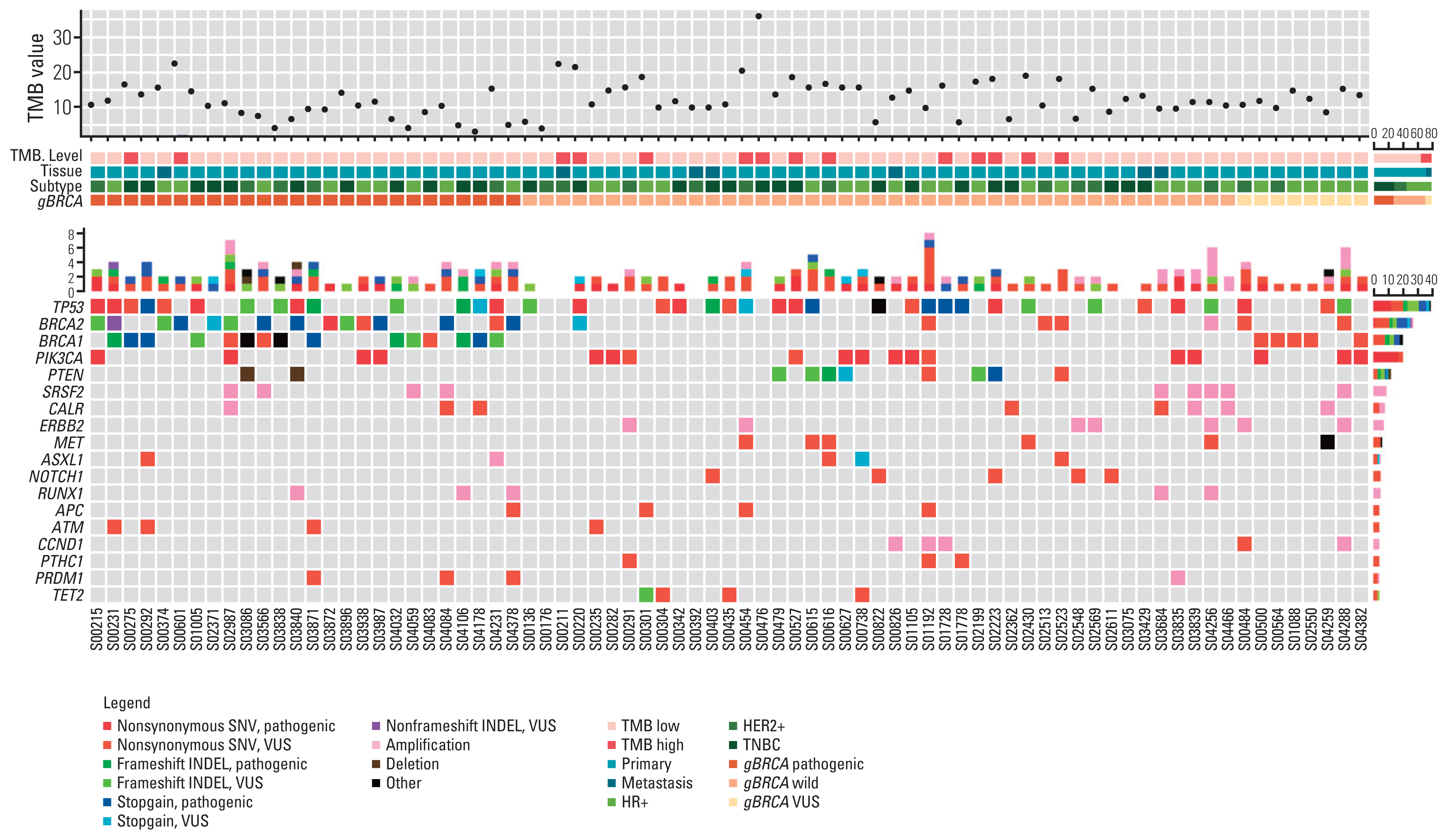
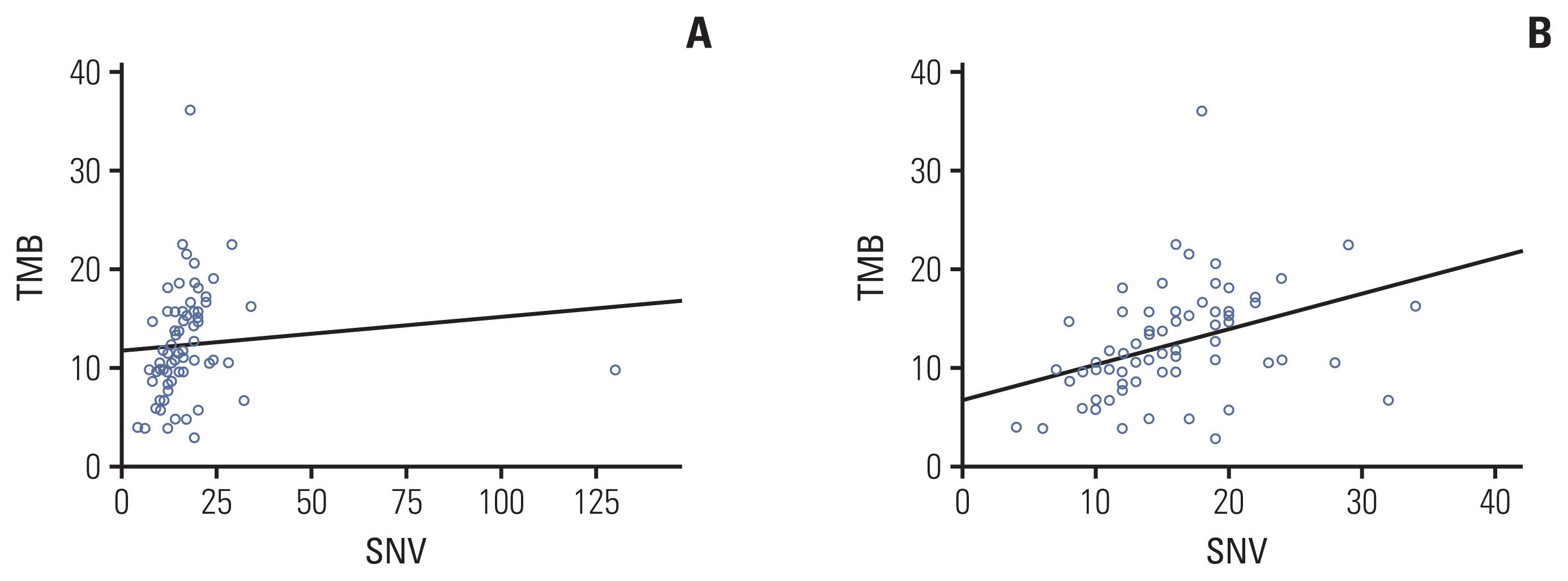
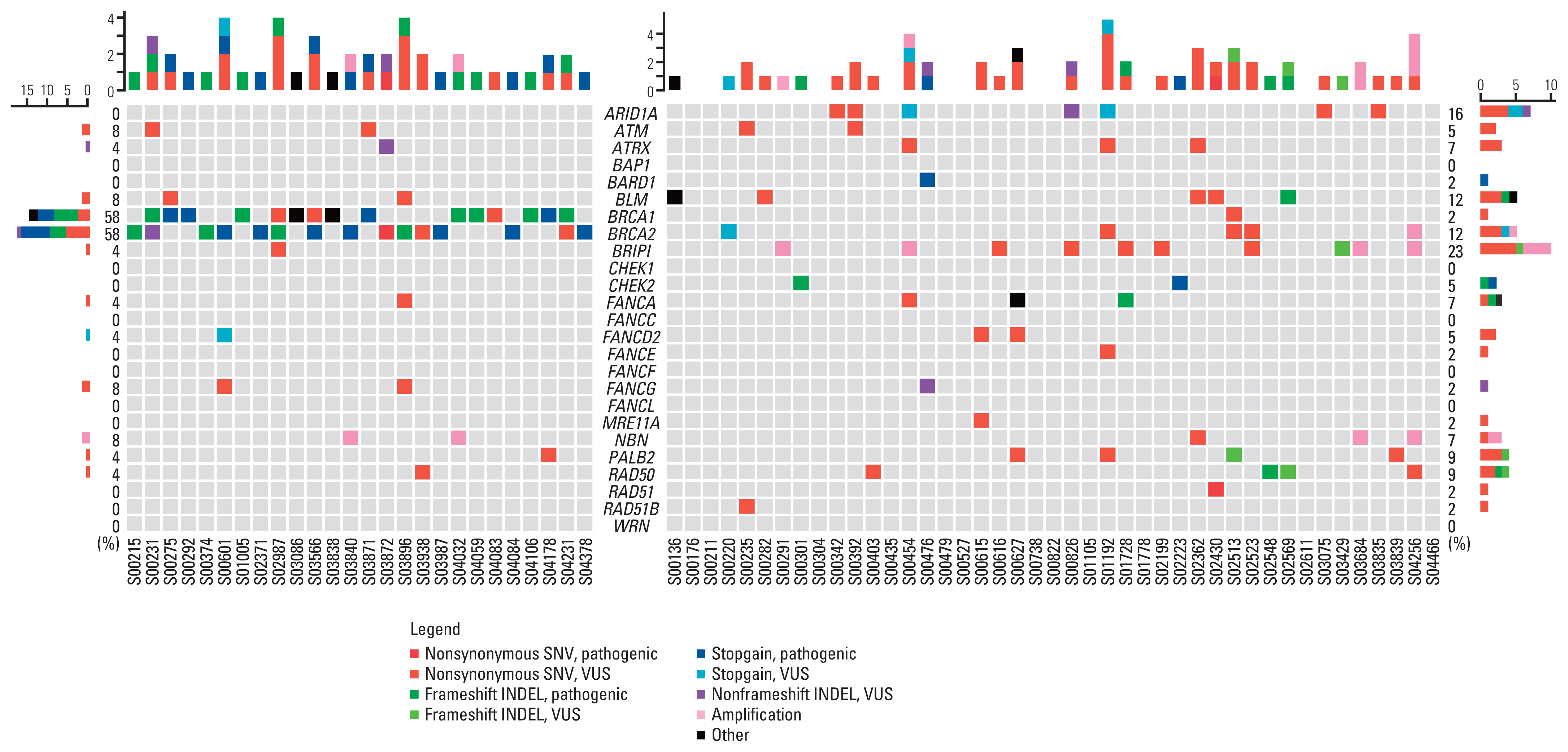
A total of 321 patients who underwent gBRCA1/2 testing were screened, and 26 patients with gBRCA1/2 pathogenic (gBRCA1/2p) variants, eight patients with gBRCA1/2 variants of uncertain significance (VUS; gBRCA1/2v), and 43 patients with gBRCA1/2 wild-type (gBRCA1/2w) were included in analysis. Mutations in TP53 (49.4%) and PIK3CA (23.4%) were frequently detected in all samples. The number of single-nucleotide variants (SNVs) per tumor tissue was higher in the gBRCA1/2w group than that in the gBRCA1/2p group (14.81 vs. 18.86, p=0.278). Tumor mutation burden (TMB) was significantly higher in the gBRCA1/2w group than in the gBRCA1/2p group (10.21 vs. 13.47, p=0.017). Except for BRCA1/2, other HR-related genes were frequently mutated in patients with gBRCA1/2w.
Conclusion
We demonstrated high sensitivity of gBRCA1/2 in tumors analyzed by NGS using a panel of tumor tissues. TMB value and aberration of non-BRCA1/2 HR-related genes differed significantly according to gBRCA1/2 pathogenicity in patients with breast cancer.
Keyword
Figure
Reference
-
References
1. Wang RF, Wang HY. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 2017; 27:11–37.2. Deng X, Nakamura Y. Cancer precision medicine: from cancer screening to drug selection and personalized immunotherapy. Trends Pharmacol Sci. 2017; 38:15–24.3. Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018; 379:753–63.4. Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017; 377:523–33.5. Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008; 26:4282–8.6. Chappuis PO, Nethercot V, Foulkes WD. Clinicopathological characteristics of BRCA1- and BRCA2-related breast cancer. Semin Surg Oncol. 2000; 18:287–95.7. Lai S, Brookes C, Prosser DO, Lan CC, Doherty E, Love DR. Diagnostic screening workflow for mutations in the BRCA1 and BRCA2 genes. Sultan Qaboos Univ Med J. 2015; 15:e58–70.8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.9. Heeke AL, Pishvaian MJ, Lynce F, Xiu J, Brody JR, Chen WJ, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018; 2018:PO.1700286.10. Hiltemann S, Jenster G, Trapman J, van der Spek P, Stubbs A. Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res. 2015; 25:1382–90.11. Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015; 7:283ra53.12. Raymond VM, Gray SW, Roychowdhury S, Joffe S, Chinnaiyan AM, Parsons DW, et al. Germline findings in tumor-only sequencing: points to consider for clinicians and laboratories. J Natl Cancer Inst. 2016; 108:djv351.13. Park S, Lee C, Ku BM, Kim M, Park WY, Kim NKD, et al. Paired analysis of tumor mutation burden calculated by targeted deep sequencing panel and whole exome sequencing in non-small cell lung cancer. BMB Rep. 2021; 54:386–91.14. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017; 9:34.15. Barroso-Sousa R, Jain E, Cohen O, Kim D, Buendia-Buendia J, Winer E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol. 2020; 31:387–94.16. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014; 371:2189–99.17. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015; 348:124–8.18. Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017; 16:2598–608.19. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019; 51:202–6.20. Choucair K, Morand S, Stanbery L, Edelman G, Dworkin L, Nemunaitis J. TMB: a promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther. 2020; 27:841–53.21. Ross JS, Gay LM, Sheehan CE, Dalvi S, Voronel O, Elvin JA, et al. Biomarkers of immune checkpoint inhibitor response in metastatic breast cancer: PD-L1 protein expression, CD274 gene amplification, and total mutational burden. J Clin Oncol. 2016; 34:3057.22. Kraya AA, Maxwell KN, Wubbenhorst B, Wenz BM, Pluta J, Rech AJ, et al. Genomic signatures predict the immunogenicity of BRCA-deficient breast cancer. Clin Cancer Res. 2019; 25:4363–74.23. Zhou Z, Li M. Evaluation of BRCA1 and BRCA2 as indicators of response to immune checkpoint inhibitors. JAMA Netw Open. 2021; 4:e217728.24. Park S, Lee E, Park S, Lee S, Nam SJ, Kim SW, et al. Clinical characteristics and exploratory genomic analyses of germline BRCA1 or BRCA2 mutations in breast cancer. Mol Cancer Res. 2020; 18:1315–25.25. Kawamoto Y, Nakamura Y, Ikeda M, Bando H, Esaki T, Ueno M, et al. Biological difference of tumour mutational burden (TMB) and microsatellite instability (MSI) status in patients (pts) with somatic vs germline BRCA1/2-mutated advanced gastrointestinal (GI) cancers using cell-free DNA (cfDNA) sequencing analysis in the GOZILA study. Ann Oncol. 2019; 30:v31–2.26. To assess safety and efficacy of agents targeting DNA damage repair with olaparib versus olaparib monotherapy. Clinical trials.gov identifier: NCT03330847 [Internet]. Bethesda, MD: U.S. National Library of Medicine;2017. [cited 2021 Apr 20]. Available from: https://clinicaltrials.gov/ct2/show/NCT033-30847 .27. Chen B, Zhang G, Li X, Ren C, Wang Y, Li K, et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging (Albany NY). 2020; 12:3140–55.28. Khoury K, Tan AR, Elliott A, Xiu J, Gatalica Z, Heeke AL, et al. Prevalence of phosphatidylinositol-3-kinase (PI3K) pathway alterations and co-alteration of other molecular markers in breast cancer. Front Oncol. 2020; 10:1475.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- BRCA1 Reversion Mutation Confers Resistance to Olaparib and Camrelizumab in a Patient with Breast Cancer Liver Metastasis
- Germline Mutations of BRCA1 Gene in Korean Breast and/or Ovarian Cancer Families
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- A Study for Germline Mutation of BRCA1 in Early Onset Breast Cancer Patients
- Novel Germline Mutations of BRCA1 and BRCA2 in Korean Familial Breast Cancer Patients