Cancer Res Treat.
2022 Oct;54(4):1209-1218. 10.4143/crt.2021.963.
Genomic Correlates of Unfavorable Outcome in Locally Advanced Cervical Cancer Treated with Neoadjuvant Chemoradiation
- Affiliations
-
- 1Cheeloo College of Medicine, Shandong University, Jinan, China
- 2Department of Radiology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
- 3Department of Gynecologic Oncology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
- 4Geneseeq Research Institute, Nanjing Geneseeq Technology Inc., Nanjing, China
- 5Department of Pathology, Shandong Cancer Hospital and Institute, Shandong First Medical University and Shandong Academy of Medical Sciences, Jinan, China
- 6School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China
- KMID: 2534200
- DOI: http://doi.org/10.4143/crt.2021.963
Abstract
- Purpose
Neoadjuvant therapy modality can increase the operability rate and mitigate pathological risks in locally advanced cervical cancer, but treatment response varies widely. It remains unclear whether genetic alterations correlate with the response to neoadjuvant therapy and disease-free survival (DFS) in locally advanced cervical cancer.
Materials and Methods
A total of 62 locally advanced cervical cancer (stage IB-IIA) patients who received neoadjuvant chemoradiation plus radical hysterectomy were retrospectively analyzed. Patients’ tumor biopsy samples were comprehensively profiled using targeted next generation sequencing. Pathologic response to neoadjuvant treatment and DFS were evaluated against the association with genomic traits.
Results
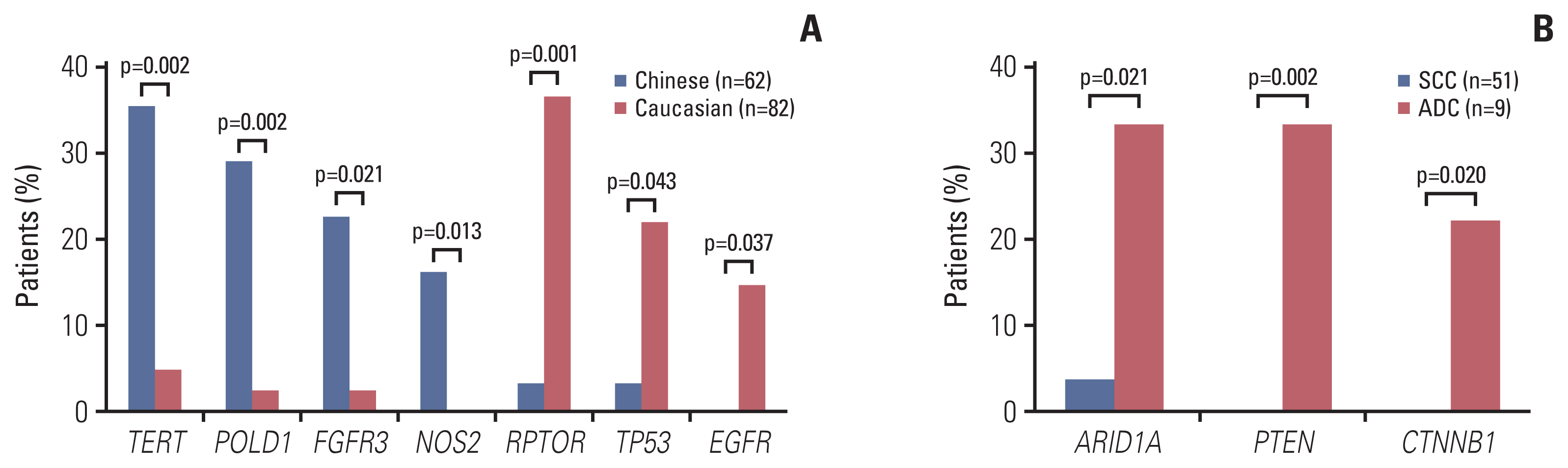
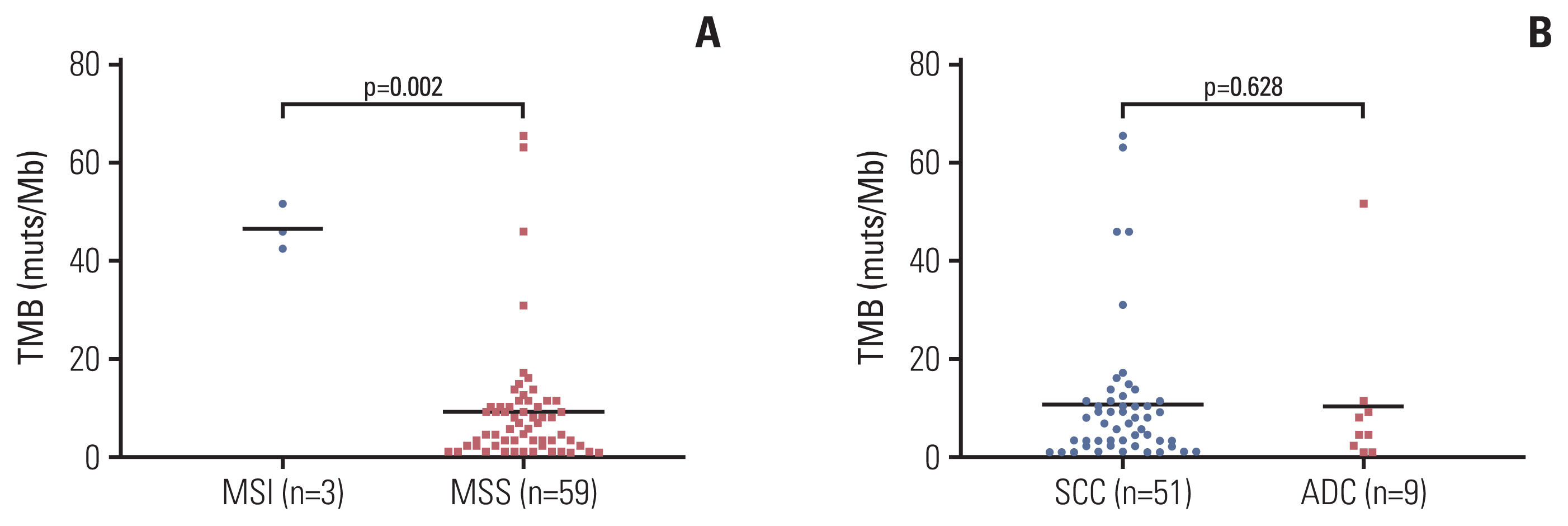
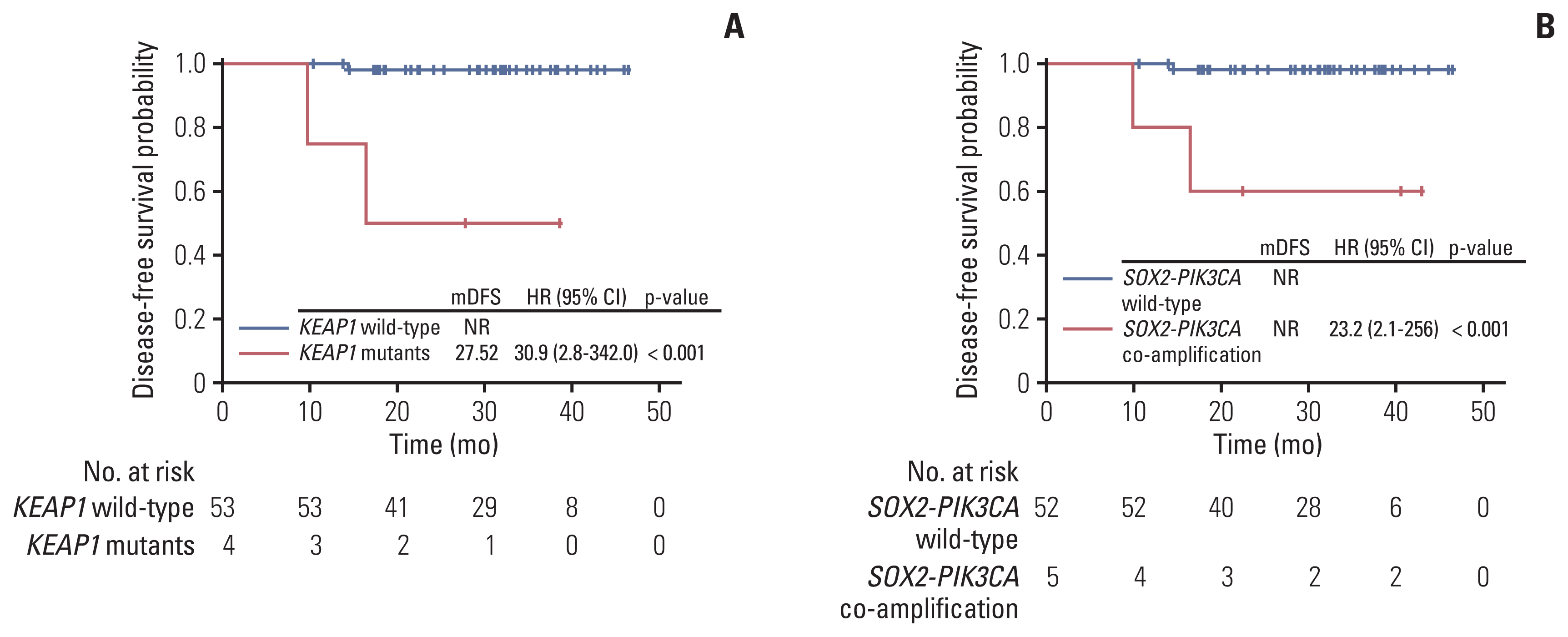
Genetic alterations of PIK3CA were most frequent (37%), comparable to that of Caucasian populations from The Cancer Genome Atlas. The mutation frequency of genes including TERT, POLD1, NOS2, and FGFR3 was significantly higher in Chinese patients whereas RPTOR, EGFR, and TP53 were underrepresented in comparison to Caucasians. Germline mutations were identified in 21% (13/62) of the cohort and more than half (57%) had mutations in DNA damage repair genes, including BRCA1/2, TP53 and PALB2. Importantly, high tumor mutation burden, TP53 polymorphism (rs1042522), and KEAP1 mutations were found to be associated with poor pathologic response to neoadjuvant chemoradiation treatment. KEAP1 mutations, PIK3CA-SOX2 co-amplification, TERC copy number gain, and TYMS polymorphism correlated with an increased risk of disease relapse.
Conclusion
We report the genomic profile of locally advanced cervical cancer patients and the distinction between Asian and Caucasian cohorts. Our findings highlight genomic traits associated with unfavorable neoadjuvant chemoradiation response and a higher risk of early disease recurrence.
Keyword
Figure
Reference
-
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.2. Small W Jr, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, et al. Cervical cancer: a global health crisis. Cancer. 2017; 123:2404–12.3. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016; 66:115–32.4. Li S, Hu T, Lv W, Zhou H, Li X, Yang R, et al. Changes in prevalence and clinical characteristics of cervical cancer in the People’s Republic of China: a study of 10,012 cases from a nationwide working group. Oncologist. 2013; 18:1101–7.5. Bahrami A, Hasanzadeh M, Shahidsales S, Farazestanian M, Hassanian SM, Moetamani Ahmadi M, et al. Genetic susceptibility in cervical cancer: From bench to bedside. J Cell Physiol. 2018; 233:1929–39.6. Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N, et al. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017; 28:iv72–83.7. Ma Y, Zhao G, Qi J, Sun P, Liu C, Qu P, et al. Neoadjuvant brachytherapy and chemotherapy followed by radical surgery for stage IB2 and IIA cervical cancer: a retrospective comparison with chemoirradiation. Mol Clin Oncol. 2018; 8:617–22.8. Peters WA 3rd, Liu PY, Barrett RJ 2nd, Stock RJ, Monk BJ, Berek JS, et al. Concurrent chemotherapy and pelvic radiation therapy compared with pelvic radiation therapy alone as adjuvant therapy after radical surgery in high-risk early-stage cancer of the cervix. J Clin Oncol. 2000; 18:1606–13.9. Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009; 105:103–4.10. Hellmann MD, Chaft JE, William WN Jr, Rusch V, Pisters KM, Kalhor N, et al. Pathological response after neoadjuvant chemotherapy in resectable non-small-cell lung cancers: proposal for the use of major pathological response as a surrogate endpoint. Lancet Oncol. 2014; 15:e42–50.11. Pataer A, Kalhor N, Correa AM, Raso MG, Erasmus JJ, Kim ES, et al. Histopathologic response criteria predict survival of patients with resected lung cancer after neoadjuvant chemotherapy. J Thorac Oncol. 2012; 7:825–32.12. Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao Y, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017; 7:583.13. Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018; 24:3097–107.14. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–20.15. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012; 22:568–76.16. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014; 30:3390–3.17. Xia H, Xue X, Ding H, Ou Q, Wu X, Nagasaka M, et al. Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: results from a large-scale survey of NTRK1 fusions in Chinese patients with lung cancer. Clin Lung Cancer. 2020; 21:247–54.18. Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016; 12:477–9.19. Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003; 348:518–27.20. Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015; 19:A68–77.21. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017; 170:605–35.22. McMurray HR, McCance DJ. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J Virol. 2003; 77:9852–61.23. Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019; 38:6172–83.24. Bertelsen B, Tuxen IV, Yde CW, Gabrielaite M, Torp MH, Kinalis S, et al. High frequency of pathogenic germline variants within homologous recombination repair in patients with advanced cancer. NPJ Genom Med. 2019; 4:13.25. Nguyen L, Martens JW, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020; 11:5584.26. Dite GS, Jenkins MA, Southey MC, Hocking JS, Giles GG, McCredie MR, et al. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003; 95:448–57.27. Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol. 2020; 38:674–85.28. Gupta S, Maheshwari A, Parab P, Mahantshetty U, Hawaldar R, Sastri Chopra S, et al. Neoadjuvant chemotherapy followed by radical surgery versus concomitant chemotherapy and radiotherapy in patients with stage IB2, IIA, or IIB squamous cervical cancer: a randomized controlled trial. J Clin Oncol. 2018; 36:1548–55.29. Kenter G, Greggi S, Vergote I, Katsaros D, Kobierski J, Massuger L, et al. Results from neoadjuvant chemotherapy followed by surgery compared to chemoradiation for stage Ib2–IIb cervical cancer, EORTC 55994. J Clin Oncol. 2019; 37(15 Suppl):5503.30. AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017; 7:818–31.31. Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 2017; 7:86–101.32. Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008; 68:1303–9.33. Voutsadakis IA. 3q26 amplifications in cervical squamous carcinomas. Curr Oncol. 2021; 28:2868–80.34. Davidson MA, Shanks EJ. 3q26–29 Amplification in head and neck squamous cell carcinoma: a review of established and prospective oncogenes. FEBS J. 2017; 284:2705–31.35. McCaughan F, Pole JC, Bankier AT, Konfortov BA, Carroll B, Falzon M, et al. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med. 2010; 182:83–91.36. Kokalj Vokac N, Cizmarevic B, Zagorac A, Zagradisnik B, Lanisnik B. An evaluation of SOX2 and hTERC gene amplifications as screening markers in oral and oropharyngeal squamous cell carcinomas. Mol Cytogenet. 2014; 7:5.37. Kawano O, Sasaki H, Okuda K, Yukiue H, Yokoyama T, Yano M, et al. PIK3CA gene amplification in Japanese non-small cell lung cancer. Lung Cancer. 2007; 58:159–60.38. Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG, Hur J, et al. PIK3CA amplification is associated with poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Oncotarget. 2016; 7:30691–701.39. Fendri A, Khabir A, Mnejja W, Sellami-Boudawara T, Daoud J, Frikha M, et al. PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma. Cancer Sci. 2009; 100:2034–9.40. Li X, Huang K, Zhang Q, Zhou J, Sun H, Tang F, et al. Genome-wide association study identifies four SNPs associated with response to platinum-based neoadjuvant chemotherapy for cervical cancer. Sci Rep. 2017; 7:41103.41. Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003; 3:387–402.42. Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti-PD-(L)1 therapies in non-small cell lung cancer. Clin Cancer Res. 2019; 25:5015–26.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Results of Radiation Alone Versus Neoadjuvant Chemotherapy and Radiation in Locally Advanced Stage of Uterine Cervical Cancer
- Recent Management of FIGO stage IB2 Cervical Cancer
- Treatment of Cervical Cancer
- The Outcome of Preoperative Chemoradiation to Locally Advanced Rectal Cancer
- Neoadjuvant Chemotherapy in Asian Patients With Locally Advanced Gastric Cancer