J Korean Med Sci.
2022 Sep;37(36):e271. 10.3346/jkms.2022.37.e271.
Efficient Segmentation for Left Atrium With Convolution Neural Network Based on Active Learning in Late Gadolinium Enhancement Magnetic Resonance Imaging
- Affiliations
-
- 1Department of Radiology, Korea University Anam Hospital, Seoul, Korea
- 2AI Center, Korea University Anam Hospital, Seoul, Korea
- 3Division of Cardiology, Department of Internal Medicine, Korea University Anam Hospital, Seoul, Korea
- 4Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- KMID: 2533226
- DOI: http://doi.org/10.3346/jkms.2022.37.e271
Abstract
- Background
To propose fully automatic segmentation of left atrium using active learning with limited dataset in late gadolinium enhancement in cardiac magnetic resonance imaging (LGE-CMRI).
Methods
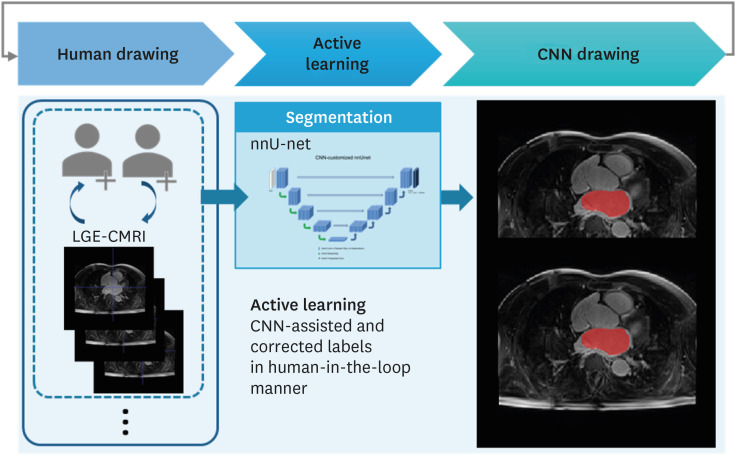
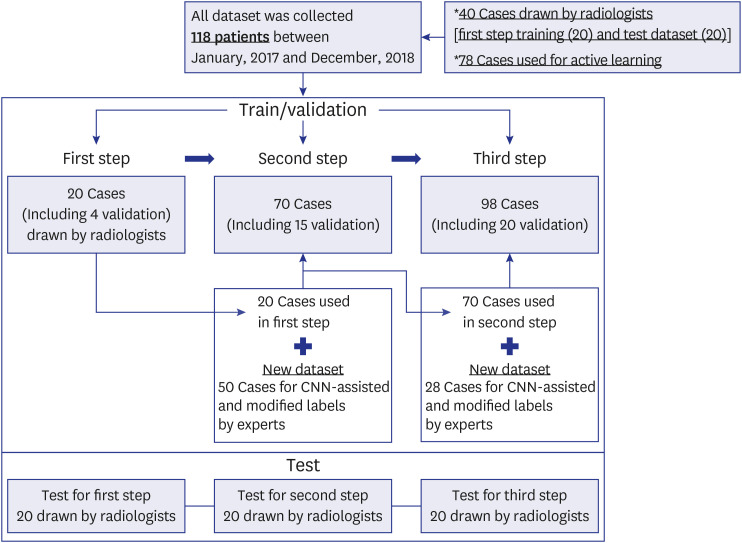
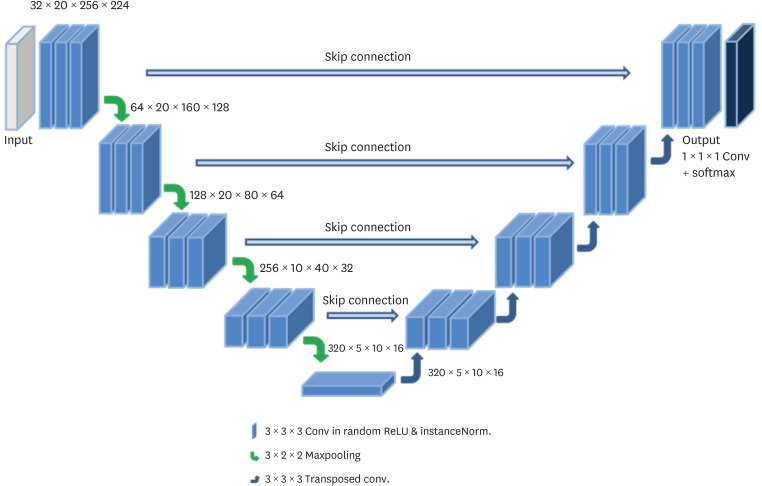
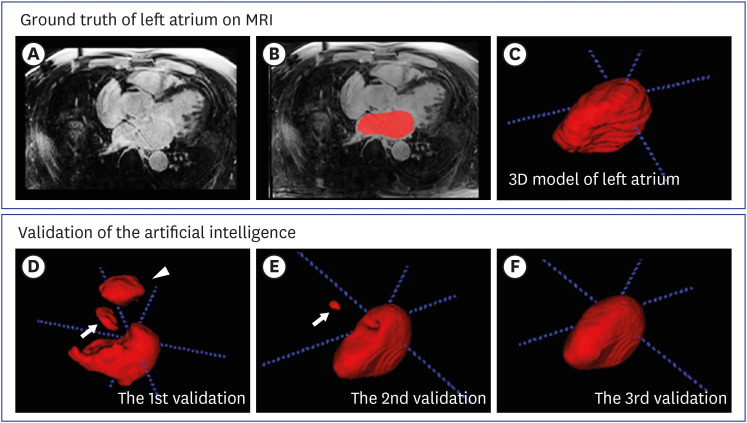
An active learning framework was developed to segment the left atrium in cardiac LGE-CMRI. Patients (n = 98) with atrial fibrillation from the Korea University Anam Hospital were enrolled. First, 20 cases were delineated for ground truths by two experts and used for training a draft model. Second, the 20 cases from the first step and 50 new cases, corrected in a human-in-the-loop manner after predicting using the draft model, were used to train the next model; all 98 cases (70 cases from the second step and 28 new cases) were trained. An additional 20 LGE-CMRI were evaluated in each step.
Results
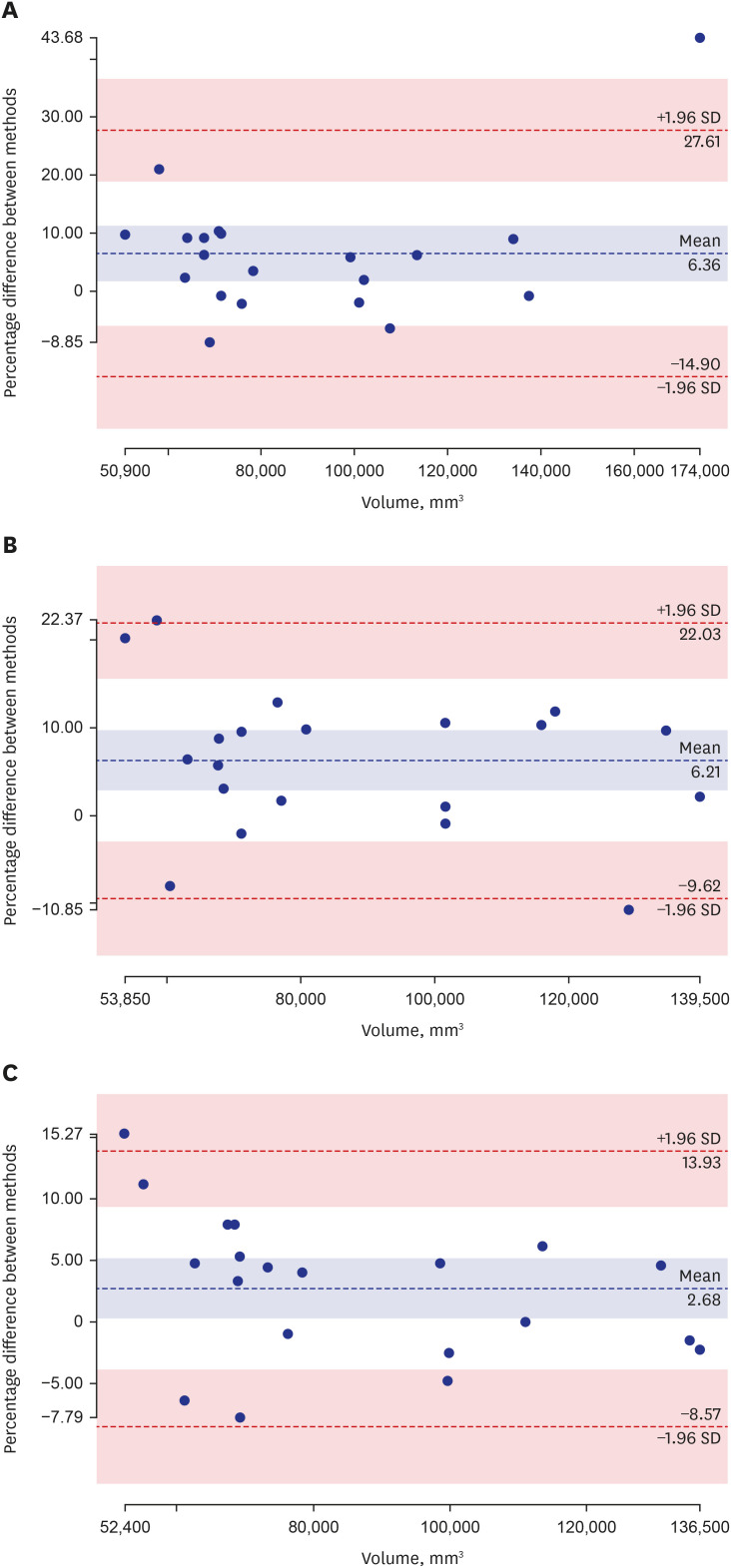
The Dice coefficients for the three steps were 0.85 ± 0.06, 0.89 ± 0.02, and 0.90 ± 0.02, respectively. The biases (95% confidence interval) in the Bland-Altman plots of each step were 6.36% (−14.90–27.61), 6.21% (−9.62–22.03), and 2.68% (−8.57–13.93). Deep active learning-based annotation times were 218 ± 31 seconds, 36.70 ± 18 seconds, and 36.56 ± 15 seconds, respectively.
Conclusion
Deep active learning reduced annotation time and enabled efficient training on limited LGE-CMRI.
Keyword
Figure
Reference
-
1. Chen H, Qi X, Cheng JZ, Heng PA. Deep contextual networks for neuronal structure segmentation. In : Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence; 2016 Feb 12–17; Phoenix, AZ, USA. Palo Alto, CA, USA: Association for the Advancement of Artificial Intelligence;2016.2. Rao M, Stough J, Chi YY, Muller K, Tracton G, Pizer SM, et al. Comparison of human and automatic segmentations of kidneys from CT images. Int J Radiat Oncol Biol Phys. 2005; 61(3):954–960. PMID: 15708280.
Article3. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In : Proceedings of MICCAI 2015: Medical Image Computing and Computer-Assisted Intervention; 2015 Oct 5–9; Munich, Germany. Berlin: Springer;2015. p. 234–241.4. Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. In : Proceedings of the 2015 IEEE International Conference on Computer Vision; 2015 Dec 7–13; Santiago, Chile. New York, NY, USA: IEEE;2015. p. 1520–1528.5. Zheng S, Jayasumana S, Romera-Paredes B, Vineet V, Su Z, Du D, et al. Conditional random fields as recurrent neural networks. In : Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV); 2015 Dec 11–18; Las Condes, Chile. New York, NY, USA: IEEE;2015. p. 1529–1537.6. Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017; 39(4):640–651. PMID: 27244717.
Article7. Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018; 40(4):834–848. PMID: 28463186.
Article8. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-net: learning dense volumetric segmentation from sparse annotation. In : Proceedings of MICCAI 2016: Medical Image Computing and Computer-Assisted Intervention; 2016 Oct 17–21; Athens, Greece. Berlin: Springer;2016. p. 424–432.9. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021; 18(2):203–211. PMID: 33288961.
Article10. Christ PF, Elshaer ME, Ettlinger F, Tatavarty S, Bickel M, Bilic P, et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In : Proceedings of MICCAI 2016: Medical Image Computing and Computer-Assisted Intervention; 2016 Oct 17–21; Athens, Greece. Berlin: Springer;2016. p. 415–423.11. Cui S, Mao L, Jiang J, Liu C, Xiong S. Automatic semantic segmentation of brain gliomas from MRI images using a deep cascaded neural network. J Healthc Eng. 2018; 2018:4940593. PMID: 29755716.
Article12. He Y, Carass A, Yun Y, Zhao C, Jedynak BM, Solomon SD, et al. Towards topological correct segmentation of macular OCT from cascaded FCNs. Fetal Infant Ophthalmic Med Image Anal (2017). 2017; 10554:202–209. PMID: 31355372.13. Roth HR, Oda H, Zhou X, Shimizu N, Yang Y, Hayashi Y, et al. An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput Med Imaging Graph. 2018; 66:90–99. PMID: 29573583.
Article14. Tang M, Zhang Z, Cobzas D, Jagersand M, Jaremko JL. Segmentation-by-detection: a cascade network for volumetric medical image segmentation. In : Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging; 2018 Apr 4–7; Washington, D.C., USA. New York, NY, USA: IEEE;2018. p. 1356–1359.15. Kasarla T, Nagendar G, Hegde G, Balasubramanian VN, Jawahar CV. (Region-based active learning for efficient labeling in semantic segmentation. In : Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV); 2019 Jan 7–11; Waikoloa Village, HI, USA. New York, NY, USA: IEEE;2019.16. Kim T, Lee KH, Ham S, Park B, Lee S, Hong D, et al. Active learning for accuracy enhancement of semantic segmentation with CNN-corrected label curations: evaluation on kidney segmentation in abdominal CT. Sci Rep. 2020; 10(1):366. PMID: 31941938.
Article17. Sourati J, Gholipour A, Dy JG, Kurugol S, Warfield SK. Active deep learning with fisher information for patch-wise semantic segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018). 2018; 11045:83–91. PMID: 30450490.
Article18. Wang G, Li W, Zuluaga MA, Pratt R, Patel PA, Aertsen M, et al. Interactive medical image segmentation using deep learning with image-specific fine tuning. IEEE Trans Med Imaging. 2018; 37(7):1562–1573. PMID: 29969407.19. Wen S, Kurc TM, Hou L, Saltz JH, Gupta RR, Batiste R, et al. Comparison of different classifiers with active learning to support quality control in nucleus segmentation in pathology images. AMIA Jt Summits Transl Sci Proc. 2018; 2017:227–236. PMID: 29888078.20. Yang L, Zhang Y, Chen J, Zhang S, Chen DZ. Suggestive annotation: a deep active learning framework for biomedical image segmentation. In : Proceedings of MICCAI 2017: Medical Image Computing and Computer-Assisted Intervention; 2017 Sep 11–13; Quebec City, QC, Canada. Berlin: Springer;2017. p. 399–407.21. Penso M, Moccia S, Scafuri S, Muscogiuri G, Pontone G, Pepi M, et al. Automated left and right ventricular chamber segmentation in cardiac magnetic resonance images using dense fully convolutional neural network. Comput Methods Programs Biomed. 2021; 204:106059. PMID: 33812305.
Article22. Zhang X, Noga M, Martin DG, Punithakumar K. Fully automated left atrium segmentation from anatomical cine long-axis MRI sequences using deep convolutional neural network with unscented Kalman filter. Med Image Anal. 2021; 68:101916. PMID: 33285484.
Article23. MONAI Consortium. MONAI architecture. Updated 2020. Accessed April 5, 2022. https://docs.monai.io/en/latest/highlights.html .
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- RE: Relation of Late Gadolinium Enhancement to Increased Ventricular Wall Stress in Dilated Cardiomyopathy
- Advanced Cardiac MR Imaging for Myocardial Characterization and Quantification: T1 Mapping
- A Self-Supervised Learning Framework for Under-Sampling Pattern Design Using Graph Convolution Network
- Convolutional Neural Network-Based Automatic Segmentation of Substantia Nigra on Nigrosome and Neuromelanin Sensitive MR Images
- Delayed Enhancement Magnetic Resonance Imaging Findings in Cardiac Amyloidosis