J Korean Med Sci.
2022 May;37(18):e144. 10.3346/jkms.2022.37.e144.
Accuracy of Cloud-Based Speech Recognition Open Application Programming Interface for Medical Terms of Korean
- Affiliations
-
- 1Rehabilitation and Prevention Center, Heart Vascular Stroke Institute, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 2Department of Biomedical Engineering, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Anesthesiology and Pain Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 4Department of Biomedical Sciences, Ajou University Graduate School of Medicine, Suwon, Korea
- 5Center for Health Promotion, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 6Interdisciplinary Program in Bioengineering, Graduate School, Seoul National University, Seoul, Korea
- 7Integrated Major in Innovative Medical Science, Seoul National University Graduate School, Seoul, Korea
- 8Institute of Medical and Biological Engineering, Medical Research Center, Seoul National University, Seoul, Korea
- KMID: 2529710
- DOI: http://doi.org/10.3346/jkms.2022.37.e144
Abstract
- Background
There are limited data on the accuracy of cloud-based speech recognition (SR) open application programming interfaces (APIs) for medical terminology. This study aimed to evaluate the medical term recognition accuracy of current available cloud-based SR open APIs in Korean.
Methods
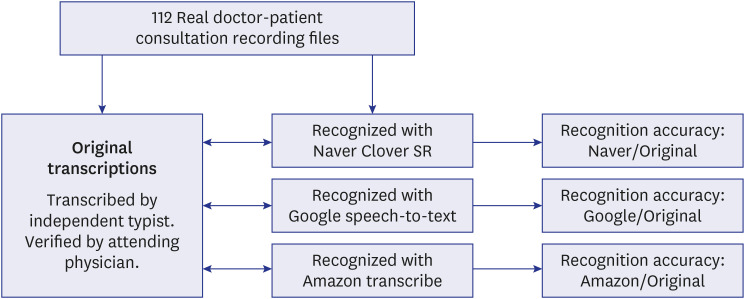
We analyzed the SR accuracy of currently available cloud-based SR open APIs using real doctor–patient conversation recordings collected from an outpatient clinic at a large tertiary medical center in Korea. For each original and SR transcription, we analyzed the accuracy rate of each cloud-based SR open API (i.e., the number of medical terms in the SR transcription per number of medical terms in the original transcription).
Results
A total of 112 doctor–patient conversation recordings were converted with three cloud-based SR open APIs (Naver Clova SR from Naver Corporation; Google Speech-toText from Alphabet Inc.; and Amazon Transcribe from Amazon), and each transcription was compared. Naver Clova SR (75.1%) showed the highest accuracy with the recognition of medical terms compared to the other open APIs (Google Speech-to-Text, 50.9%, P < 0.001; Amazon Transcribe, 57.9%, P < 0.001), and Amazon Transcribe demonstrated higher recognition accuracy compared to Google Speech-to-Text (P< 0.001). In the sub-analysis, Naver Clova SR showed the highest accuracy in all areas according to word classes, but the accuracy of words longer than five characters showed no statistical differences (Naver Clova SR, 52.6%; Google Speech-to-Text, 56.3%; Amazon Transcribe, 36.6%).
Conclusion
Among three current cloud-based SR open APIs, Naver Clova SR which manufactured by Korean company showed highest accuracy of medical terms in Korean, compared to Google Speech-to-Text and Amazon Transcribe. Although limitations are existing in the recognition of medical terminology, there is a lot of rooms for improvement of this promising technology by combining strengths of each SR engines.
Figure
Reference
-
1. Hodgson T, Coiera E. Risks and benefits of speech recognition for clinical documentation: a systematic review. J Am Med Inform Assoc. 2016; 23(e1):e169–e179. PMID: 26578226.2. Zhou L, Blackley SV, Kowalski L, Doan R, Acker WW, Landman AB, et al. Analysis of errors in dictated clinical documents assisted by speech recognition software and professional transcriptionists. JAMA Netw Open. 2018; 1(3):e180530. PMID: 30370424.3. Hinton G, Deng L, Yu D, Dahl G, Mohamed A, Jaitly N, et al. Deep neural networks for acoustic modeling in speech recognition: the shared views of four research groups. IEEE Signal Process Mag. 2012; 29(6):82–97.4. Chiu CC, Sainath TN, Wu Y, Prabhavalkar R, Nguyen P, Chen Z, et al. State-of-the-art speech recognition with sequence-to-sequence models. In : Proceeding of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2018 April 15–20; Calgary, Canada. Manhattan, NY, USA: IEEE;2018. p. 4774–4778.5. Johnson M, Lapkin S, Long V, Sanchez P, Suominen H, Basilakis J, et al. A systematic review of speech recognition technology in health care. BMC Med Inform Decis Mak. 2014; 14:94. PMID: 25351845.6. Yoo HJ, Seo S, Im SW, Gim GY. The performance evaluation of continuous speech recognition based on Korean phonological rules of cloud-based speech recognition open API. Int J Networked Distrib Comput. 2021; 9(1):10–18.7. Spinazze P, Aardoom J, Chavannes N, Kasteleyn M. The computer will see you now: overcoming barriers to adoption of computer-assisted history taking (CAHT) in primary care. J Med Internet Res. 2021; 23(2):e19306. PMID: 33625360.8. Elmore N, Burt J, Abel G, Maratos FA, Montague J, Campbell J, et al. Investigating the relationship between consultation length and patient experience: a cross-sectional study in primary care. Br J Gen Pract. 2016; 66(653):e896–e903. PMID: 27777231.9. Sinsky C, Colligan L, Li L, Prgomet M, Reynolds S, Goeders L, et al. Allocation of physician time in ambulatory practice: a time and motion study in 4 specialties. Ann Intern Med. 2016; 165(11):753–760. PMID: 27595430.10. Basma S, Lord B, Jacks LM, Rizk M, Scaranelo AM. Error rates in breast imaging reports: comparison of automatic speech recognition and dictation transcription. AJR Am J Roentgenol. 2011; 197(4):923–927. PMID: 21940580.11. Kim D, Oh J, Im H, Yoon M, Park J, Lee J. Automatic classification of the Korean triage acuity scale in simulated emergency rooms using speech recognition and natural language processing: a proof of concept study. J Korean Med Sci. 2021; 36(27):e175. PMID: 34254471.12. Kim S, Lee J, Choi SG, Ji S, Kang J, Kim J, et al. Building a Korean conversational speech database in the emergency medical domain. Phon Speech Sci. 2020; 12(4):81–90.13. Kauppinen T, Koivikko MP, Ahovuo J. Improvement of report workflow and productivity using speech recognition--a follow-up study. J Digit Imaging. 2008; 21(4):378–382. PMID: 18437491.14. Blackley BV, Schubert VD, Goss FR, Al Assad W, Garabedian PM, Zhou L. Physician use of speech recognition versus typing in clinical documentation: a controlled observational study. Int J Med Inform. 2020; 141:104178. PMID: 32521449.15. Ghatnekar S, Faletsky A, Nambudiri VE. Digital scribe utility and barriers to implementation in clinical practice: a scoping review. Health Technol (Berl). 2021; 11(4):803–809. PMID: 34094806.16. Swayamsiddha S, Prashant K, Shaw D, Mohanty C. The prospective of artificial intelligence in COVID-19 pandemic. Health Technol (Berl). 2021; 11(6):1311–1320. PMID: 34603925.17. Choi SJ, Kim JB. Comparison analysis of speech recognition open APIs’ accuracy. Asia Pac J Multimed Serv Converg Art Humanit Sociol. 2017; 7(8):411–418.18. Kaushal A, Altman R, Langlotz C. Geographic distribution of US cohorts used to train deep learning algorithms. JAMA. 2020; 324(12):1212–1213. PMID: 32960230.19. Amazon. Guide of Amazon Transcribe Medical. Updated 2022. Accessed January 18, 2022. https://aws.amazon.com/ko/transcribe/medical/ .20. Choi DH, Park IN, Shin M, Kim EG, Shin DR. Korean erroneous sentence classification with Integrated Eojeol Embedding. IEEE Access. 2021; 9:81778–81785.21. Leslie D, Mazumder A, Peppin A, Wolters MK, Hagerty A. Does “AI” stand for augmenting inequality in the era of COVID-19 healthcare? BMJ. 2021; 372:n304. PMID: 33722847.22. Aminololama-Shakeri S, López JE. The doctor-patient relationship with artificial intelligence. AJR Am J Roentgenol. 2019; 212(2):308–310. PMID: 30540210.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Institutional Applications of Eclipse Scripting Programming Interface to Clinical Workflows in Radiation Oncology
- Correction System of a Mis-recognized Medical Vocabulary of Speech-based Electronic Medical Record
- Classification of dental implant systems using cloud-based deep learning algorithm: an experimental study
- Evaluation of Speech Perception Abilities in Children with Cochlear Implants
- Introduction to an Open Source Internet-Based Testing Program for Medical Student Examinations