Obstet Gynecol Sci.
2022 Jan;65(1):52-63. 10.5468/ogs.21237.
Signature of arylacetamide deacetylase expression is associated with prognosis and immune infiltration in ovarian cancer
- Affiliations
-
- 1Department of Obstetrics and Gynecology, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- KMID: 2524685
- DOI: http://doi.org/10.5468/ogs.21237
Abstract
Objective
The role of the protein-coding gene arylacetamide deacetylase (AADAC) in the prognostication of ovarian cancer remains uncertain. We aimed to identify and validate its prognostic value using integrated bioinformatics analyses.
Methods
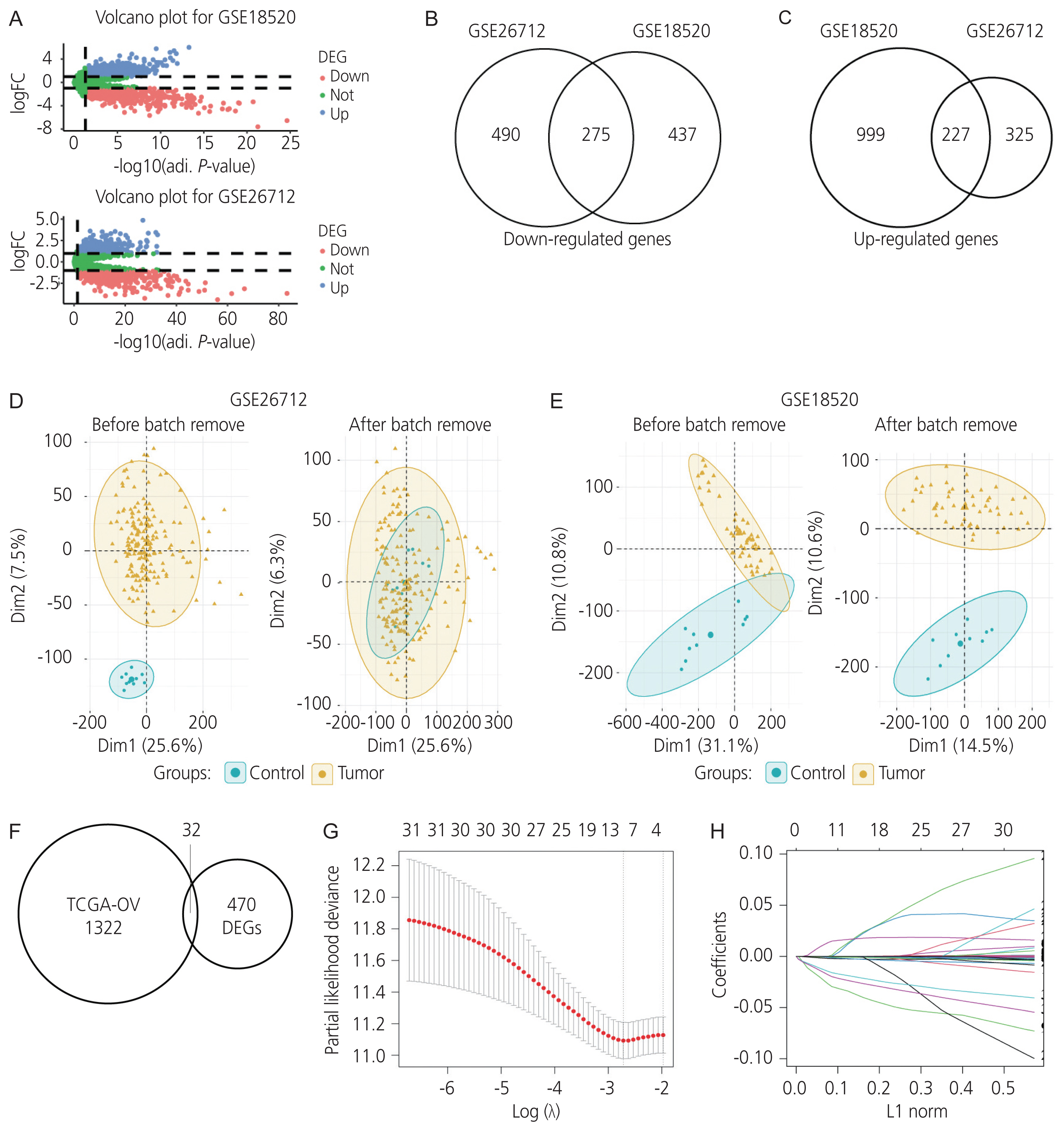
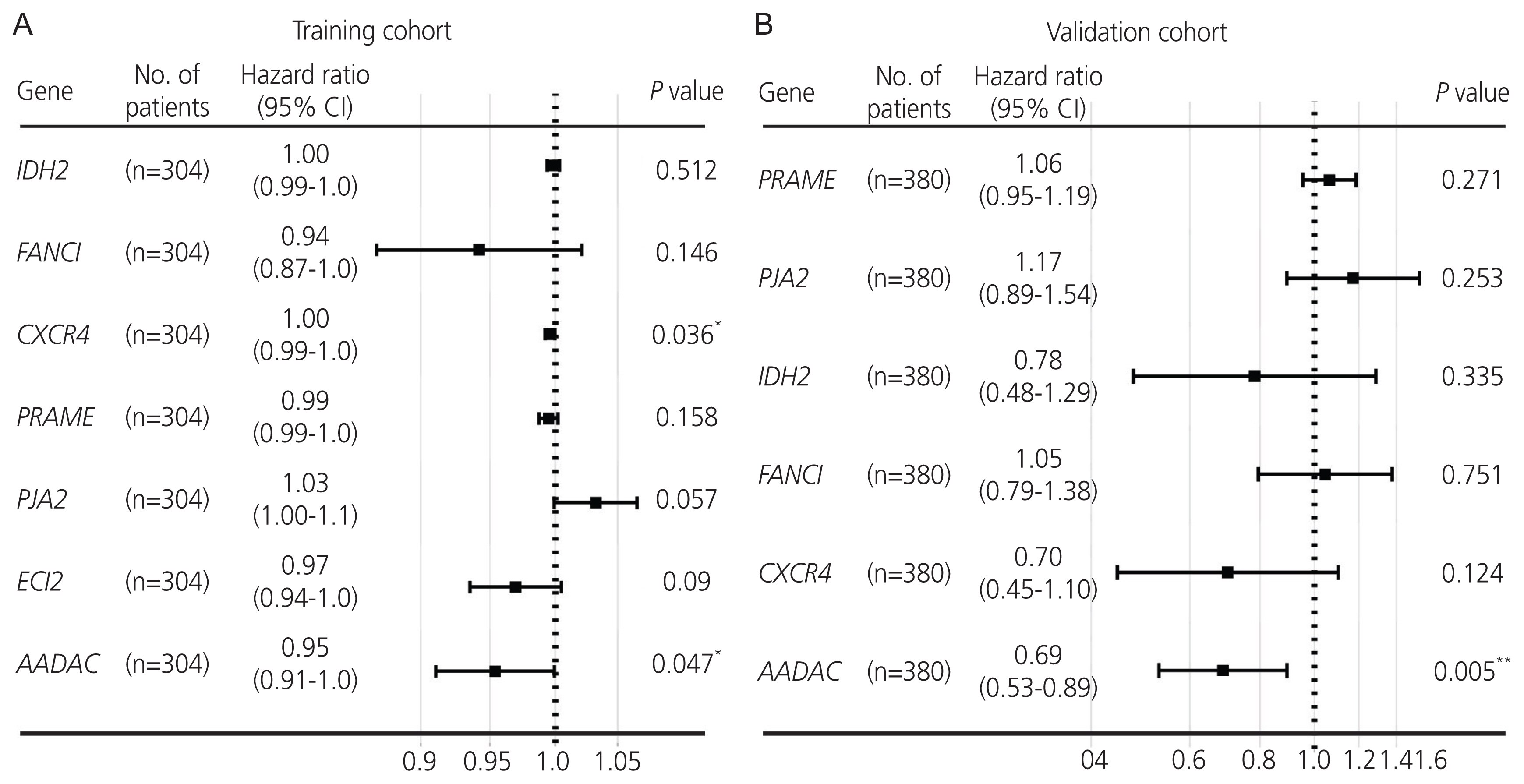
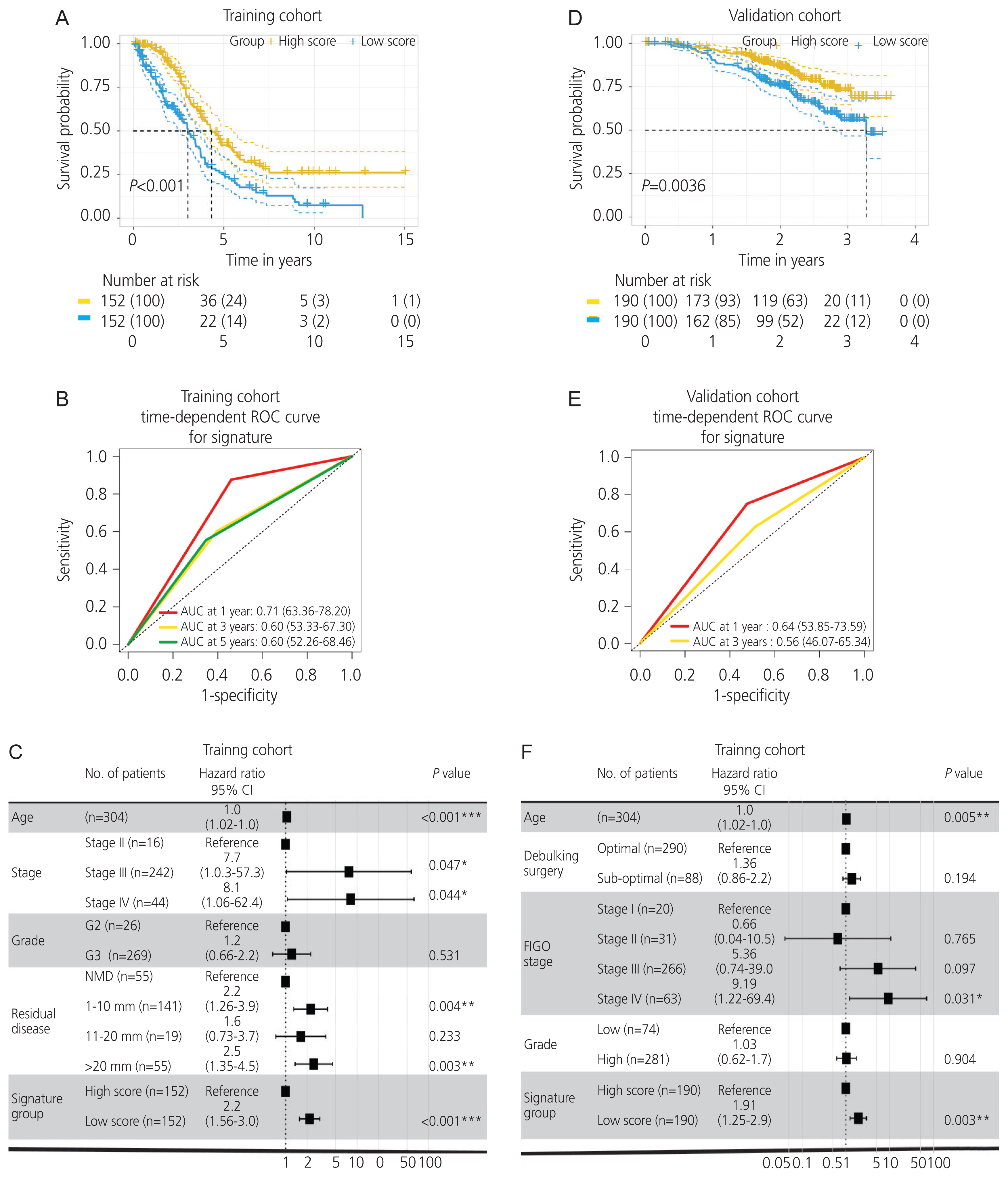
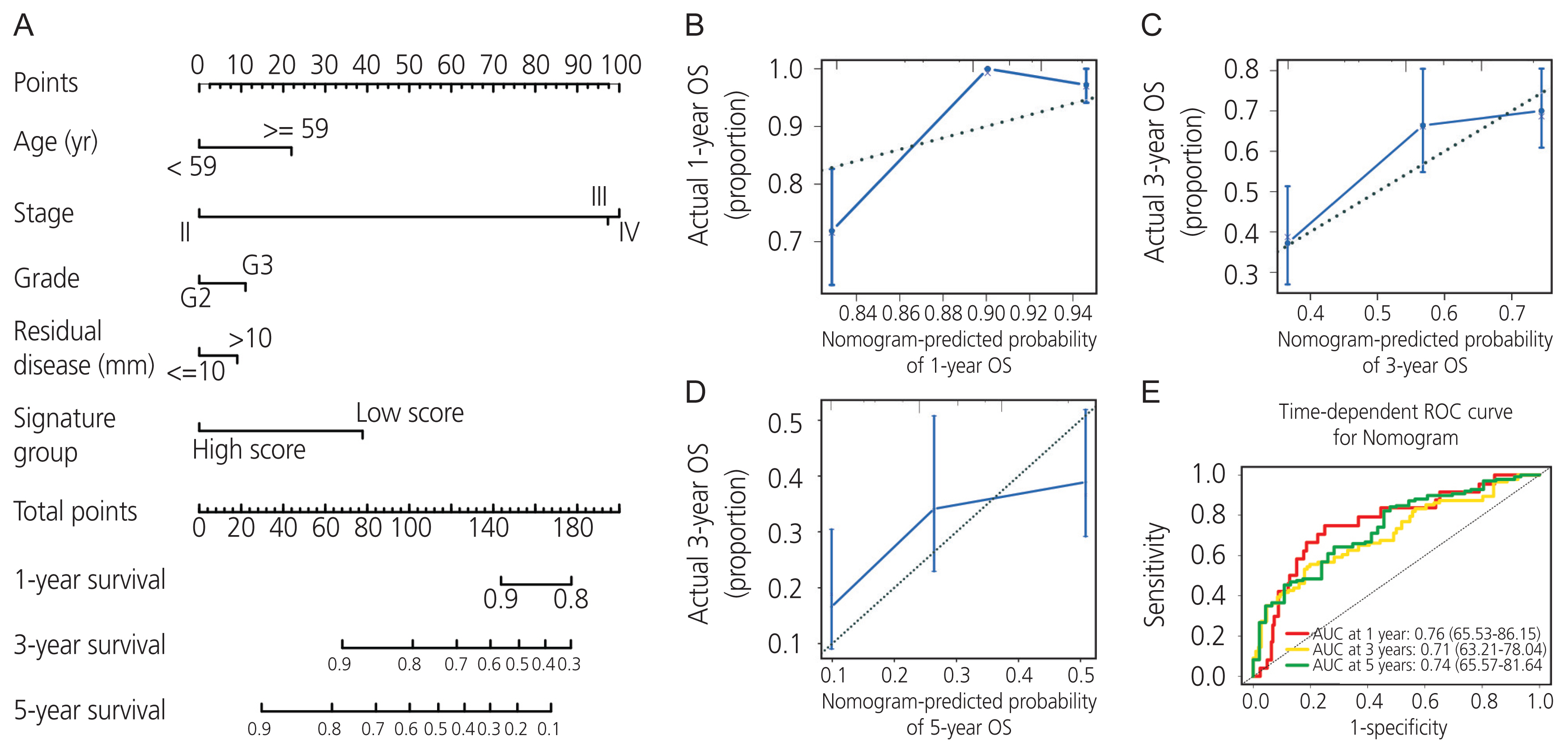
Gene expression profiles of RNA-sequencing and microarray data were retrieved from The Cancer Genome Atlas and Gene Expression Omnibus. Univariate and multivariate Cox regression models were used to evaluate the prognostic value of gene expression. The predictive accuracy of the gene signature model was evaluated using a time-dependent receiver operating characteristic (ROC) curve. In addition, the correlation between immune infiltration and AADAC was identified. A nomogram of the gene signature with clinical parameters was constructed to estimate the clinical application of the signature for survival prediction in patients with ovarian cancer.
Results
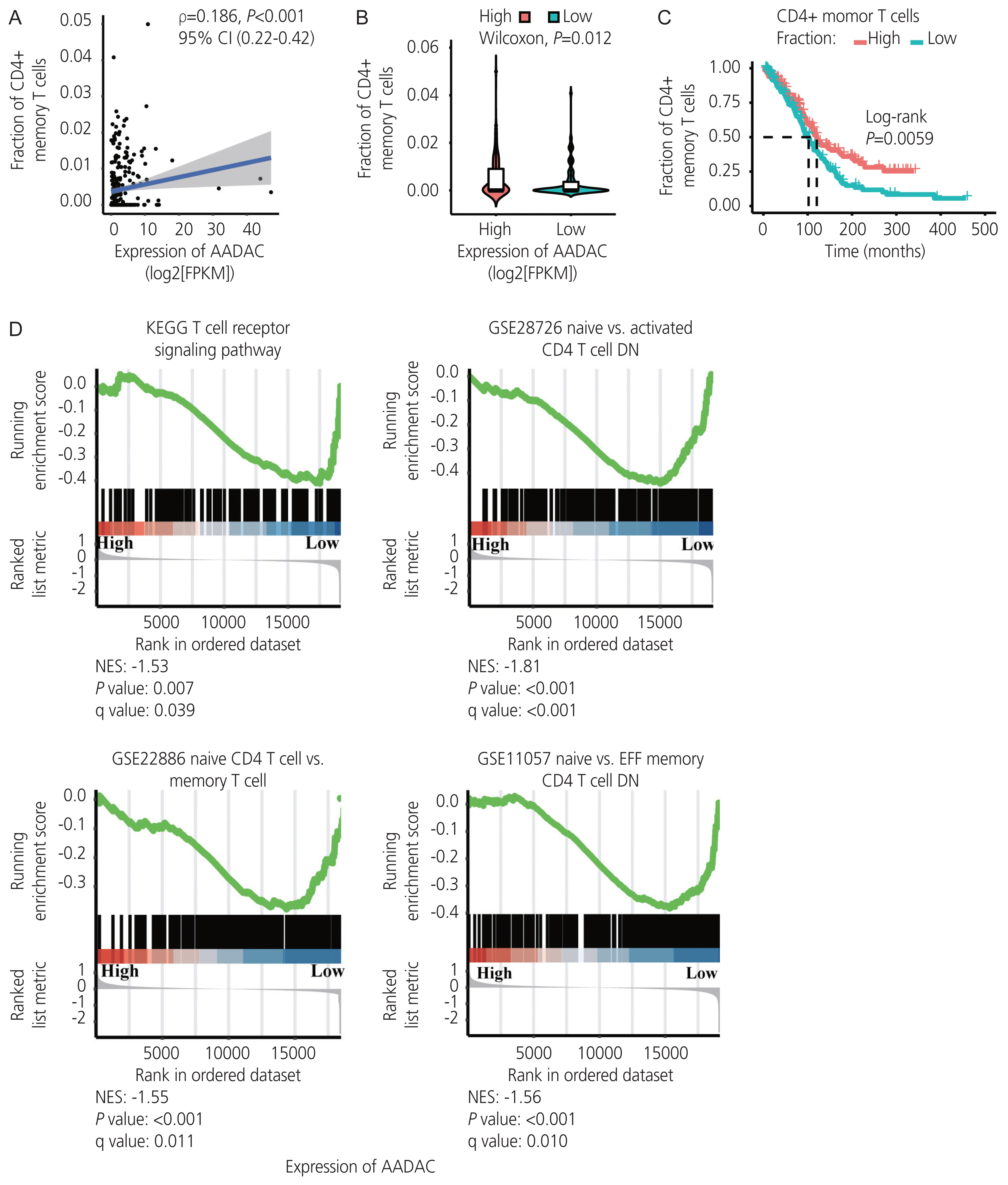
Univariate and multivariate Cox regression analyses in the training and validation cohorts indicated that a high AADAC expression signature was significantly and independently correlated with better survival outcomes in ovarian cancer. AADAC upregulation positively correlated with the infiltration of CD4+ memory T cells. Immunological signature gene sets were significantly enriched in CD4+ T cell regulation pathways. The area under the curve of the time-dependent ROC for overall survival indicated that the constructed nomogram had a moderate predictive ability for prognostic prediction in ovarian cancer.
Conclusion
AADAC expression signature significantly and independently correlated with the survival outcome and CD4+ memory T cell infiltration in ovarian cancer, indicating its potential applicability in the prediction of prognosis and immunotherapy efficacy.
Keyword
Figure
Reference
-
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.
Article2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020; 70:7–30.
Article3. Probst MR, Beer M, Beer D, Jenö P, Meyer UA, Gasser R. Human liver arylacetamide deacetylase. Molecular cloning of a novel esterase involved in the metabolic activation of arylamine carcinogens with high sequence similarity to hormone-sensitive lipase. J Biol Chem. 1994; 269:21650–6.
Article4. Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform. 2011; 12:449–62.
Article5. Kobayashi Y, Fukami T, Nakajima A, Watanabe A, Nakajima M, Yokoi T. Species differences in tissue distribution and enzyme activities of arylacetamide deacetylase in human, rat, and mouse. Drug Metab Dispos. 2012; 40:671–9.
Article6. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014; 13:397–406.
Article7. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020; 38:675–8.
Article8. Kauffmann A, Gentleman R, Huber W. array Quality Metrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009; 25:415–6.9. Jeffrey TL, Johnson WE, Hilary SP, Elana JF, Andrew EJ, Yuqing Z, et al. Surrogate Variable Analysis [Internet]. Bioconductor. c2020. [cited 2020 Oct 17]. Available from: https://www.bioconductor.org/packages/3.11/bioc/html/sva.html .10. Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008; 25:1–18.11. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47.
Article12. Heagerty PJ. Heagerty and packaging by Paramita Saha-Chaudhuri. survivalROC: Time-dependent ROC curve estimation from censored survival data [Internet]. CRAN;c2013. [cited 2013 Jan 13]. Available from: https://cran.rproject.org/web/packages/survivalROC/index.html .13. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017; 18:220.
Article14. Therneau TM, Lumley T, Elizabeth A, Cynthia C. survival: Survival Analysis [Internet]. CRAN;c2021. [cited 2021 Mar 16]. Available from: https://mran.microsoft.com/web/packages/survival/index.html .15. Harrell FE Jr. rms: Regression Modeling Strategies [Internet]. CRAN;c2021. [cited 2021 Feb 06]. Available from: https://CRAN.R-project.org/package=rms .16. Berek JS, Kehoe ST, Kumar L, Friedlander M. Cancer of the ovary, fallopian tube, and peritoneum. Int J Gynaecol Obstet. 2018; 143(Suppl 2):59–78.
Article17. Tan TZ, Miow QH, Huang RY, Wong MK, Ye J, Lau JA, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO MOL MED. 2013; 5:1051–66.
Article18. Calura E, Ciciani M, Sambugaro A, Paracchini L, Benvenuto G, Milite S, et al. Transcriptional characterization of stage I epithelial ovarian cancer: a multicentric study. Cells. 2019; 8:1554.
Article19. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–15.20. Fukami T, Iida A, Konishi K, Nakajima M. Human arylacetamide deacetylase hydrolyzes ketoconazole to trigger hepatocellular toxicity. Biochem Pharmacol. 2016; 116:153–61.
Article21. Jiang J, Chen X, Zhong D. Arylacetamide deacetylase is involved in vicagrel bioactivation in humans. Front Pharmacol. 2017; 8:846.
Article22. Zhang N, Jiang J, Tang S, Sun G. Redictive value of neutrophil-lymphocyte ratio and platelet-lymphocyte ratio in non-small cell lung cancer patients treated with immune checkpoint inhibitors: a meta-analysis. Int Immunopharmacol. 2020; 85:106677.23. Chon S, Lee S, Jeong D, Lim S, Lee K, Shin J. Elevated platelet lymphocyte ratio is a poor prognostic factor in advanced epithelial ovarian cancer. J Gynecol Obstet Hum Reprod. 2021; 50:101849.
Article24. Wang H, Ding Y, Li N, Wu L, Gao Y, Xiao C, et al. Prognostic value of neutrophil-lymphocyte ratio, platelet-lymphocyte ratio, and combined neutrophil-lymphocyte ratio and platelet-lymphocyte ratio in stage IV advanced gastric cancer. Front Oncol. 2020; 10:841.
Article25. Toyohara T, Roudnicky F, Florido MHC, Nakano T, Yu H, Katsuki S, et al. Patient hiPSCs identify vascular smooth muscle arylacetamide deacetylase as protective against atherosclerosis. Cell Stem Cell. 2020; 27:178–80.
Article26. Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni M, et al. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet. 2018; 9:265.
Article27. Zhang AW, McPherson A, Milne K, Kroeger DR, Hamilton PT, Miranda A, et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell. 2018; 173:1755–69e22.
Article28. Jiménez-Sánchez A, Cybulska P, Mager KL, Koplev S, Cast O, Couturier DL, et al. Unraveling tumor-immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat Genet. 2020; 52:582–93.
Article29. Odunsi K. Immunotherapy in ovarian cancer. Ann Oncol. 2017; 28(suppl_8):viii1–7.
Article30. Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003; 348:203–13.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development and validation of a stromal-immune signature to predict prognosis in intrahepatic cholangiocarcinoma
- SLAMF7 predicts prognosis and correlates with immune infiltration in serous ovarian carcinoma
- Identification and validation of a signature based on myofibroblastic cancer-associated fibroblast marker genes for predicting prognosis, immune infiltration, and therapeutic response in bladder cancer
- Identification of an immune-related risk signature and nomogram predicting the overall survival in patients with endometrial cancer
- Correspondence to editorial on “Development and validation of a stromal-immune signature to predict prognosis in intrahepatic cholangiocarcinoma”