Healthc Inform Res.
2021 Jul;27(3):214-221. 10.4258/hir.2021.27.3.214.
Machine Learning for Antibiotic Resistance Prediction: A Prototype Using Off-the-Shelf Techniques and Entry-Level Data to Guide Empiric Antimicrobial Therapy
- Affiliations
-
- 1School of Science and Technology, Hellenic Open University, Patras, Greece
- 2IT Department, Sismanogleio General Hospital, Marousi, Greece
- 3Independent Department of Quality Control, Research and Continuing Education, Sismanogleio General Hospital, Marousi, Greece
- 4Intensive Care Unit, Sismanogleio General Hospital, Marousi, Greece
- 5Clinical Microbiology Laboratory, Sismanogleio General Hospital, Marousi, Greece
- 6Internal Medicine Department, Sismanogleio General Hospital, Marousi, Greece
- 7Hematology Laboratory, Sismanogleio General Hospital, Marousi, Greece
- KMID: 2519037
- DOI: http://doi.org/10.4258/hir.2021.27.3.214
Abstract
Objectives
In the era of increasing antimicrobial resistance, the need for early identification and prompt treatment of multi-drug-resistant infections is crucial for achieving favorable outcomes in critically ill patients. As traditional microbiological susceptibility testing requires at least 24 hours, automated machine learning (AutoML) techniques could be used as clinical decision support tools to predict antimicrobial resistance and select appropriate empirical antibiotic treatment.
Methods
An antimicrobial susceptibility dataset of 11,496 instances from 499 patients admitted to the internal medicine wards of a public hospital in Greece was processed by using Microsoft Azure AutoML to evaluate antibiotic susceptibility predictions using patients’ simple demographic characteristics, as well as previous antibiotic susceptibility testing, without any concomitant clinical data. Furthermore, the balanced dataset was also processed using the same procedure. The datasets contained the attributes of sex, age, sample type, Gram stain, 44 antimicrobial substances, and the antibiotic susceptibility results.
Results
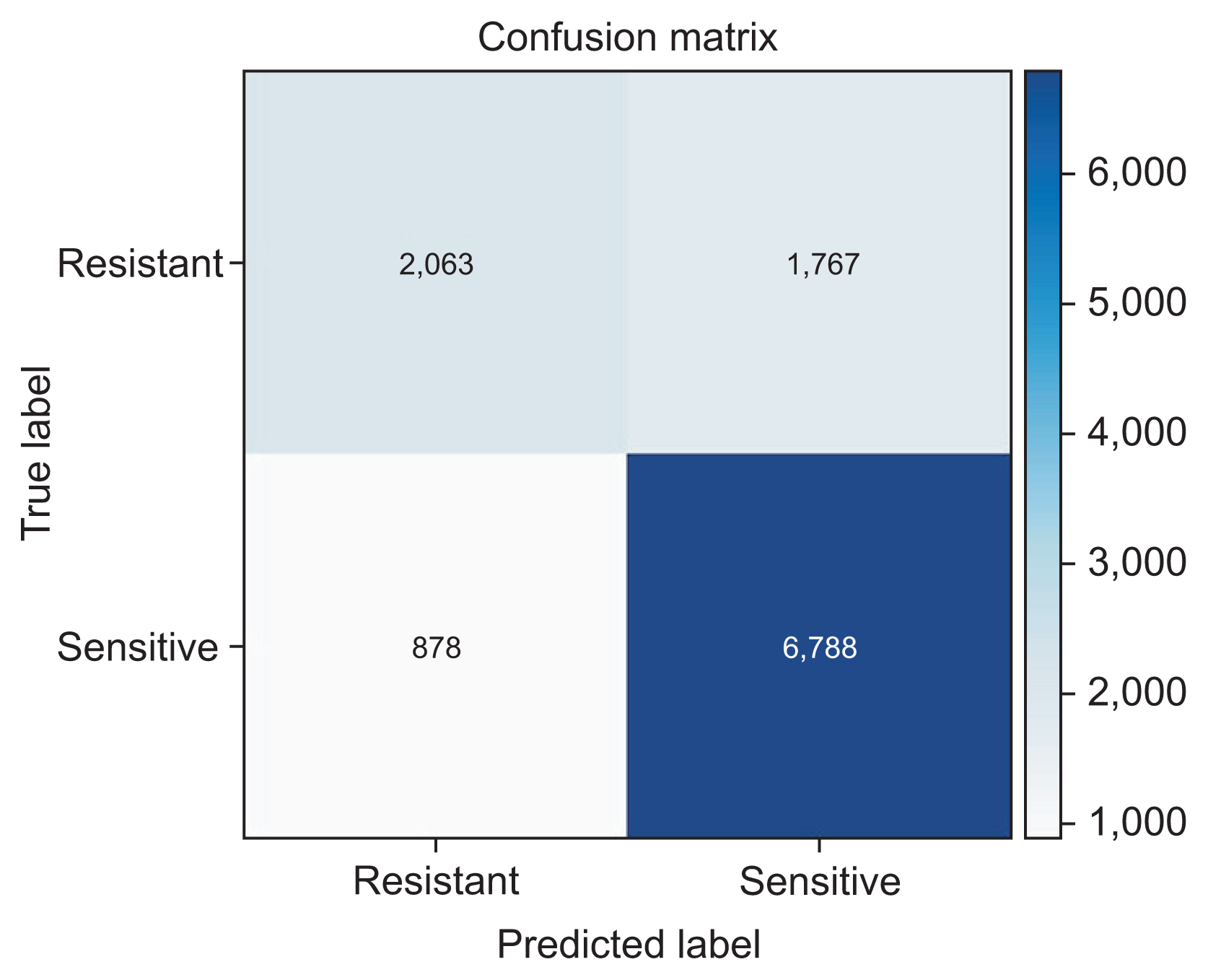
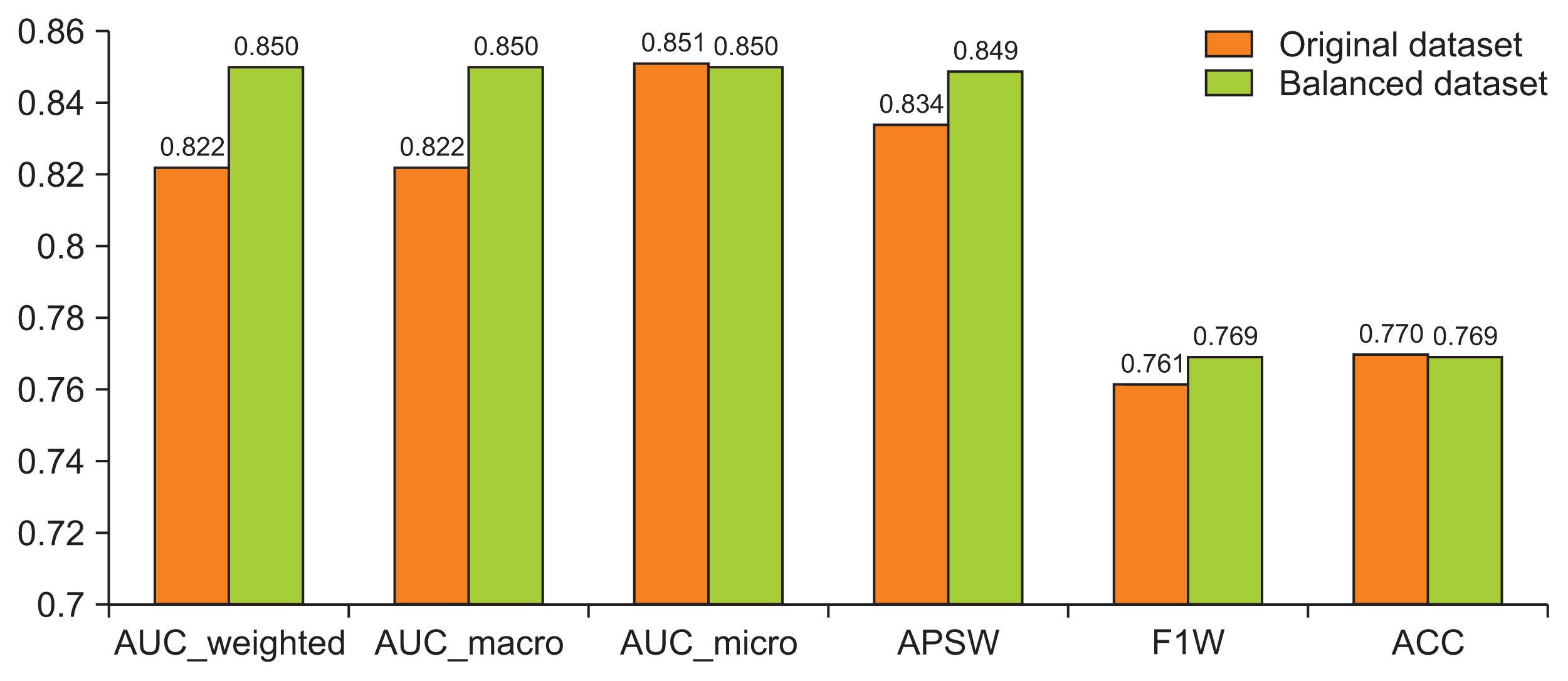
The stack ensemble technique achieved the best results in the original and balanced dataset with an area under the curve-weighted metric of 0.822 and 0.850, respectively.
Conclusions
Implementation of AutoML for antimicrobial susceptibility data can provide clinicians useful information regarding possible antibiotic resistance and aid them in selecting appropriate empirical antibiotic therapy by taking into consideration the local antimicrobial resistance ecosystem.
Figure
Reference
-
References
1. Gandra S, Barter DM, Laxminarayan R. Economic burden of antibiotic resistance: how much do we really know? Clin Microbiol Infect. 2014; 20(10):973–80.
Article2. World Health Organization. No time to wait: securing the future from drug-resistant infections [Internet]. Geneva, Switzerland: World Health Organization;2019. [cited at 2021 Jul 27]. Available from: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections .3. Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015; 45(6):568–85.4. Feretzakis G, Loupelis E, Sakagianni A, Skarmoutsou N, Michelidou S, Velentza A, et al. A 2-year single-centre audit on antibiotic resistance of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae strains from an intensive care unit and other wards in a general public hospital in Greece. Antibiotics (Basel). 2019; 8(2):62.
Article5. Feretzakis G, Loupelis E, Petropoulou S, Christopoulos C, Lada M, Martsoukou M, et al. Using microbiological data analysis to tackle antibiotic resistance of Klebsiella pneumoniae. Mantas J, Hasman A, Gallos P, Kolokathi A, Househ MS, Liaskos J, editors. Health informatics vision: from data via information to knowledge. Amsterdam, The Netherlands: IOS Press;2019. p. 180–3.6. Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, et al. Diagnosis and treatment of adults with community-acquired pneumonia: an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019; 200(7):e45–e67.
Article7. McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020; 577(7788):89–94.
Article8. Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019; 6(2):94–8.
Article9. Waring J, Lindvall C, Umeton R. Automated machine learning: review of the state-of-the-art and opportunities for healthcare. Artif Intell Med. 2020; 104:101822.
Article10. Feretzakis G, Loupelis E, Sakagianni A, Kalles D, Martsoukou M, Lada M, et al. Using machine learning techniques to aid empirical antibiotic therapy decisions in the intensive care unit of a general hospital in Greece. Antibiotics (Basel). 2020; 9(2):50.
Article11. Martinez-Aguero S, Mora-Jimenez I, Lerida-Garcia J, Alvarez-Rodriguez J, Soguero-Ruiz C. Machine learning techniques to identify antimicrobial resistance in the intensive care unit. Entropy (Basel). 2019; 21(6):603.12. Oonsivilai M, Mo Y, Luangasanatip N, Lubell Y, Miliya T, Tan P, et al. Using machine learning to guide targeted and locally-tailored empiric antibiotic prescribing in a children’s hospital in Cambodia. Wellcome Open Res. 2018; 3:131.
Article13. MacFadden DR, Coburn B, Shah N, Robicsek A, Savage R, Elligsen M, et al. Decision-support models for empiric antibiotic selection in Gram-negative bloodstream infections. Clin Microbiol Infect. 2019; 25(1):108.e1–108.e7.
Article14. Kolozsvari LR, Konya J, Paget J, Schellevis FG, Sandor J, Szollosi GJ, et al. Patient-related factors, antibiotic prescribing and antimicrobial resistance of the commensal Staphylococcus aureus and Streptococcus pneumoniae in a healthy population: Hungarian results of the APRES study. BMC Infect Dis. 2019; 19(1):253.
Article15. Ben-Ami R, Rodriguez-Bano J, Arslan H, Pitout JD, Quentin C, Calbo ES, et al. A multinational survey of risk factors for infection with extended-spectrum beta-lactamase-producing enterobacteriaceae in nonhospitalized patients. Clin Infect Dis. 2009; 49(5):682–90.16. Set up AutoML training with Python [Internet]. Redmond (WA): Microsoft;2021. [cited at 2021 Jul 28]. Available from: https://docs.microsoft.com/en-us/azure/machine-learning/how-to-configure-auto-train .17. Bengio Y, Grandvalet Y. No unbiased estimator of the variance of k-fold cross-validation. J Mach Learn Res. 2004; 5:1089–105.18. SMOTE [Internet]. Redmond (WA): Microsoft;2019. [cited at 2021 Jul 28]. Available from: https://docs.microsoft.com/en-us/azure/machine-learning/algorithm-module-reference/smote .19. Evaluate automated machine learning experiment results [Internet]. Redmond (WA): Microsoft;2020. [cited at 2021 Jul 28]. Available from: https://docs.microsoft.com/en-us/azure/machine-learning/how-to-understand-automated-ml .20. Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006; 27(8):861–74.
Article21. Sewell M. Ensemble learning. London, UK: University College London;2011.22. Zhang C, Ma Y. Ensemble machine learning: methods and applications. New York (NY): Springer Science & Business Media;2012.23. Feretzakis G, Loupelis E, Sakagianni A, Kalles D, Lada M, Christopoulos C, et al. Using machine learning algorithms to predict antimicrobial resistance and assist empirical treatment. Stud Health Technol Inform. 2020; 272:75–8.24. Cohen G, Hilario M, Sax H, Hugonnet S. Asymmetrical margin approach to surveillance of nosocomial infections using support vector classification. In : Proceedings of the Intelligent Data Analysis in Medicine and Pharmacology; 2003 Oct 19ă22; Protaras, Cyprus.25. Cohen G, Hilario M, Sax H, Hugonnet S, Pellegrini C, Geissbuhler A. An application of one-class support vector machine to nosocomial infection detection. Stud Health Technol Inform. 2004; 107(Pt 1):716–20.26. Cohen G, Hilario M, Sax H, Hugonnet S, Geissbuhler A. Learning from imbalanced data in surveillance of nosocomial infection. Artif Intell Med. 2006; 37(1):7–18.
Article27. He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009; 21(9):1263–84.28. Blagus R, Lusa L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinformatics. 2013; 14:106.
Article29. World Health Organization. Molecular methods for antimicrobial resistance (AMR) diagnostics to enhance the Global Antimicrobial Resistance Surveillance System. Geneva, Switzerland: World Health Organization;2019.30. Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect. 2017; 23(1):2–22.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Machine Learning vs. Statistical Model for Prediction Modelling: Application in Medical Imaging Research
- Prediction of Diabetic Neuropathy Using Machine Learning Techniques
- Application of Machine Learning in Rhinology: A State of the Art Review
- Predictive Modeling of Outcomes After Traumatic and Nontraumatic Spinal Cord Injury Using Machine Learning: Review of Current Progress and Future Directions
- Prediction of dental caries in 12-year-old children using machine-learning algorithms