J Liver Cancer.
2021 Mar;21(1):1-11. 10.17998/jlc.21.1.1.
Ras Mitogen-activated Protein Kinase Signaling and Kinase Suppressor of Ras as Therapeutic Targets for Hepatocellular Carcinoma
- Affiliations
-
- 1Department of Genetic Engineering, Kyung Hee University College of Life Sciences, Yongin, Korea
- KMID: 2514230
- DOI: http://doi.org/10.17998/jlc.21.1.1
Abstract
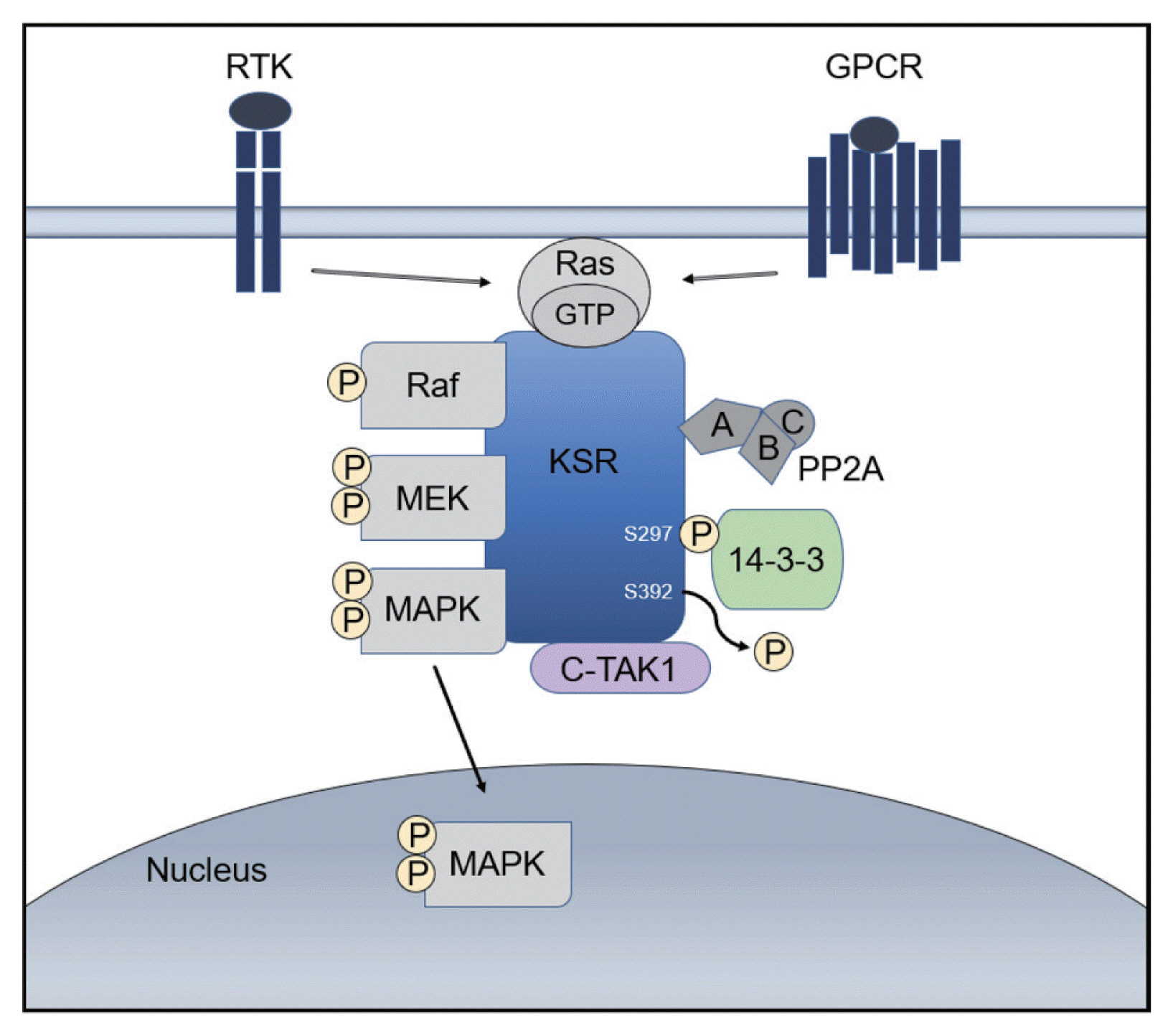
- Hepatocellular carcinoma (HCC) is a high incidence cancer and a major health concern worldwide. Among the many molecular signaling pathways that are dysregulated in HCC, the Ras mitogen-activated protein kinase (Ras/Raf/MAPK) signaling pathway has gained renewed attention from basic and clinical researchers. Mutations in Ras and Raf genes which are known to activate the Ras/Raf/MAPK signaling pathway have been infrequently detected in human HCC; however, the Ras/Raf/MAPK signaling pathway is activated in more than 50% of HCC cases, suggesting an alternative mechanism for the activation of the signaling pathway. Kinase suppressor of Ras acts as a molecular scaffold for facilitating the assembly of Ras/Raf/MAPK signaling pathway components and has been implicated in the regulation of this signaling pathway. In this review, we provide important insights into the cellular and molecular mechanisms involved in the activation of the Ras/Raf/MAPK signaling pathway and discuss potential therapeutic strategies for HCC.
Keyword
Figure
Reference
-
1. Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013; 47(Suppl):S2–S6.2. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010; 7:448–458.3. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018; 391:1301–1314.4. Knudsen ES, Gopal P, Singal AG. The changing landscape of hepatocellular carcinoma: etiology, genetics, and therapy. Am J Pathol. 2014; 184:574–583.5. Gnoni A, Licchetta A, Memeo R, Argentiero A, Solimando AG, Longo V, et al. Role of BRAF in hepatocellular carcinoma: a rationale for future targeted cancer therapies. Medicina (Kaunas). 2019; 55:754.6. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003; 362:1907–1917.7. Quetglas IM, Moeini A, Pinyol R, Llovet JM. Integration of genomic information in the clinical management of HCC. Best Pract Res Clin Gastroenterol. 2014; 28:831–842.8. Liu M, Jiang L, Guan XY. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014; 5:673–691.9. Dimri M, Satyanarayana A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers (Basel). 2020; 12:491.10. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008; 359:378–390.11. Caruso S, Calatayud AL, Pilet J, La Bella T, Rekik S, Imbeaud S, et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology. 2019; 157:760–776.12. Galuppo R, Ramaiah D, Ponte OM, Gedaly R. Molecular therapies in hepatocellular carcinoma: what can we target? Dig Dis Sci. 2014; 59:1688–1697.13. Chen C, Wang G. Mechanisms of hepatocellular carcinoma and challenges and opportunities for molecular targeted therapy. World J Hepatol. 2015; 7:1964–1970.14. Ito Y, Sasaki Y, Horimoto M, Wada S, Tanaka Y, Kasahara A, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998; 27:951–958.15. Delire B, Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest. 2015; 45:609–623.16. Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, et al. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003; 17:1263–1293.17. Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007; 1773:1177–1195.18. Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007; 26:3279–3290.19. Razidlo GL, Kortum RL, Haferbier JL, Lewis RE. Phosphorylation regulates KSR1 stability, ERK activation, and cell proliferation. J Biol Chem. 2004; 279:47808–47814.20. Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995; 83:903–913.21. Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995; 83:889–901.22. Ory S, Zhou M, Conrads TP, Veenstra TD, Morrison DK. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr Biol. 2003; 13:1356–1364.23. Neilsen BK, Frodyma DE, Lewis RE, Fisher KW. KSR as a therapeutic target for Ras-dependent cancers. Expert Opin Ther Targets. 2017; 21:499–509.24. Frodyma D, Neilsen B, Costanzo-Garvey D, Fisher K, Lewis R. Coordinating ERK signaling via the molecular scaffold kinase suppressor of Ras. F1000Res. 2017; 6:1621.25. Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995; 83:879–888.26. Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 2002; 16:427–438.27. Clapéron A, Therrien M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene. 2007; 26:3143–3158.28. McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci U S A. 2009; 106:11022–11027.29. Koveal D, Schuh-Nuhfer N, Ritt D, Page R, Morrison DK, Peti W. A CC-SAM, for coiled coil-sterile α motif, domain targets the scaffold KSR-1 to specific sites in the plasma membrane. Sci Signal. 2012; 5:ra94.30. Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005; 6:827–837.31. Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, et al. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci U S A. 1997; 94:12792–12796.32. Costanzo-Garvey DL, Pfluger PT, Dougherty MK, Stock JL, Boehm M, Chaika O, et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009; 10:366–378.33. Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR). J Mol Biol. 2002; 315:435–446.34. Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, et al. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999; 19:229–240.35. Morrison DK. KSR: a MAPK scaffold of the Ras pathway? J Cell Sci. 2001; 114(Pt 9):1609–1612.36. Nguyen A, Burack WR, Stock JL, Kortum R, Chaika OV, Afkarian M, et al. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol Cell Biol. 2002; 22:3035–3045.37. Ritt DA, Daar IO, Morrison DK. KSR regulation of the Raf-MEK-ERK cascade. Methods Enzymol. 2006; 407:224–237.38. Lozano J, Xing R, Cai Z, Jensen HL, Trempus C, Mark W, et al. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003; 63:4232–4238.39. Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol. 2004; 24:4407–4416.40. McKay MM, Freeman AK, Morrison DK. Complexity in KSR function revealed by Raf inhibitor and KSR structure studies. Small GTPases. 2011; 2:276–281.41. Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015; 12:408–424.42. Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011; 2:344–358.43. Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012; 380:358–365.44. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011; 364:2507–2516.45. Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012; 367:1694–1703.46. Taketomi A, Shirabe K, Muto J, Yoshiya S, Motomura T, Mano Y, et al. A rare point mutation in the Ras oncogene in hepatocellular carcinoma. Surg Today. 2013; 43:289–292.47. Harada N, Oshima H, Katoh M, Tamai Y, Oshima M, Taketo MM. Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 2004; 64:48–54.48. Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006; 130:1117–1128.49. Chen L, Shi Y, Jiang CY, Wei LX, Wang YL, Dai GH. Expression and prognostic role of pan-Ras, Raf-1, pMEK1 and pERK1/2 in patients with hepatocellular carcinoma. Eur J Surg Oncol. 2011; 37:513–520.50. Neuzillet C, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. MEK in cancer and cancer therapy. Pharmacol Ther. 2014; 141:160–171.51. Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology. 2000; 32:958–961.52. Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, et al. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013; 9:e1003248.53. Min L, He B, Hui L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin Cancer Biol. 2011; 21:10–20.54. Roovers K, Assoian RK. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays. 2000; 22:818–826.55. Gysin S, Lee SH, Dean NM, McMahon M. Pharmacologic inhibition of RAF-->MEK-->ERK signaling elicits pancreatic cancer cell cycle arrest through induced expression of p27Kip1. Cancer Res. 2005; 65:4870–4880.56. Maurer G, Tarkowski B, Baccarini M. Raf kinases in cancer-roles and therapeutic opportunities. Oncogene. 2011; 30:3477–3488.57. Ballif BA, Blenis J. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. 2001; 12:397–408.58. Tan X, Tamori Y, Egami H, Ishikawa S, Kurizaki T, Takai E, et al. Analysis of invasion-metastasis mechanism in pancreatic cancer: involvement of tight junction transmembrane protein occludin and MEK/ERK signal transduction pathway in cancer cell dissociation. Oncol Rep. 2004; 11:993–998.59. Tan X, Egami H, Abe M, Nozawa F, Hirota M, Ogawa M. Involvement of MMP-7 in invasion of pancreatic cancer cells through activation of the EGFR mediated MEK-ERK signal transduction pathway. J Clin Pathol. 2005; 58:1242–1248.60. Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta. 2010; 1802:396–405.61. Zhang M, Ma Q, Hu H, Zhang D, Li J, Ma G, et al. Stem cell factor/c-kit signaling enhances invasion of pancreatic cancer cells via HIF-1α under normoxic condition. Cancer Lett. 2011; 303:108–117.62. Dhawan NS, Scopton AP, Dar AC. Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature. 2016; 537:112–116.63. Fisher KW, Das B, Kim HS, Clymer BK, Gehring D, Smith DR, et al. AMPK promotes aberrant PGC1β expression to support human colon tumor cell survival. Mol Cell Biol. 2015; 35:3866–3879.64. Kortum RL, Fernandez MR, Costanzo-Garvey DL, Johnson HJ, Fisher KW, Volle DJ, et al. Caveolin-1 is required for kinase suppressor of Ras 1 (KSR1)-mediated extracellular signal-regulated kinase 1/2 activation, H-RasV12-induced senescence, and transformation. Mol Cell Biol. 2014; 34:3461–3472.65. Koral K, Paranjpe S, Bowen WC, Mars W, Luo J, Michalopoulos GK. Leukocyte-specific protein 1: a novel regulator of hepatocellular proliferation and migration deleted in human hepatocellular carcinoma. Hepatology. 2015; 61:537–547.66. Jindal A, Thadi A, Shailubhai K. Hepatocellular carcinoma: etiology and current and future drugs. J Clin Exp Hepatol. 2019; 9:221–232.67. Hojjat-Farsangi M. Small-molecule inhibitors of the receptor tyrosine kinases: promising tools for targeted cancer therapies. Int J Mol Sci. 2014; 15:13768–13801.68. Garuti L, Roberti M, Bottegoni G. Non-ATP competitive protein kinase inhibitors. Curr Med Chem. 2010; 17:2804–2821.69. Coriat R, Nicco C, Chéreau C, Mir O, Alexandre J, Ropert S, et al. Sorafenib-induced hepatocellular carcinoma cell death depends on reactive oxygen species production in vitro and in vivo. Mol Cancer Ther. 2012; 11:2284–2293.70. Zhang HL, Zhu Y, Qin XJ, Wang CF, Yao XD, Zhang SL, et al. c-KIT: potential predictive factor for the efficacy of sorafenib in metastatic renal cell carcinoma with sarcomatoid feature. Clin Genitourin Cancer. 2013; 11:134–140.71. Schmieder R, Puehler F, Neuhaus R, Kissel M, Adjei AA, Miner JN, et al. Allosteric MEK1/2 inhibitor refametinib (BAY 86-9766) in combination with sorafenib exhibits antitumor activity in preclinical murine and rat models of hepatocellular carcinoma. Neoplasia. 2013; 15:1161–1171.72. Huether A, Hopfner M, Sutter AP, Baradari V, Schuppan D, Scherubl H. Signaling pathways involved in the inhibition of epidermal growth factor receptor by erlotinib in hepatocellular cancer. World J Gastroenterol. 2006; 12:5160–5167.73. Philip PA, Mahoney MR, Allmer C, Thomas J, Pitot HC, Kim G, et al. Phase II study of Erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J Clin Oncol. 2005; 23:6657–6663.74. Cheng AL, Kang YK, Lin DY, Park JW, Kudo M, Qin S, et al. Sunitinib versus sorafenib in advanced hepatocellular cancer: results of a randomized phase III trial. J Clin Oncol. 2013; 31:4067–4075.75. Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019; 13:125–137.76. Bruix J, Tak WY, Gasbarrini A, Santoro A, Colombo M, Lim HY, et al. Regorafenib as second-line therapy for intermediate or advanced hepatocellular carcinoma: multicentre, open-label, phase II safety study. Eur J Cancer. 2013; 49:3412–3419.77. Tai WT, Chu PY, Shiau CW, Chen YL, Li YS, Hung MH, et al. STAT3 mediates regorafenib-induced apoptosis in hepatocellular carcinoma. Clin Cancer Res. 2014; 20:5768–5776.78. Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017; 389:56–66.79. Yamamoto Y, Matsui J, Matsushima T, Obaishi H, Miyazaki K, Nakamura K, et al. Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vasc Cell. 2014; 6:18.80. Nakazawa Y, Kawano S, Matsui J, Funahashi Y, Tohyama O, Muto H, et al. Multitargeting strategy using lenvatinib and golvatinib: maximizing anti-angiogenesis activity in a preclinical cancer model. Cancer Sci. 2015; 106:201–207.81. Cochin V, Gross-Goupil M, Ravaud A, Godbert Y, Le Moulec S. Cabozantinib: Mechanism of action, efficacy and indications. Bull Cancer. 2017; 104:393–401.82. Schöffski P, Gordon M, Smith DC, Kurzrock R, Daud A, Vogelzang NJ, et al. Phase II randomised discontinuation trial of cabozantinib in patients with advanced solid tumours. Eur J Cancer. 2017; 86:296–304.83. McCall JL, Gehring D, Clymer BK, Fisher KW, Das B, Kelly DL, et al. KSR1 and EPHB4 regulate Myc and PGC1β to promote survival of human colon tumors. Mol Cell Biol. 2016; 36:2246–2261.84. Zhang J, Zafrullah M, Yang X, Yin X, Zhang Z, Fuks Z, et al. Down-regulation of KSR1 in pancreatic cancer xenografts by antisense oligonucleotide correlates with tumor drug uptake. Cancer Biol Ther. 2008; 7:1490–1495.85. Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009; 461:542–545.86. Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011; 472:366–369.87. Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002; 297:63–64.88. Ng IO, Guan XY, Poon RT, Fan ST, Lee JM. Determination of the molecular relationship between multiple tumour nodules in hepatocellular carcinoma differentiates multicentric origin from intrahepatic metastasis. J Pathol. 2003; 199:345–353.89. Tao Y, Ruan J, Yeh SH, Lu X, Wang Y, Zhai W, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proc Natl Acad Sci U S A. 2011; 108:12042–12047.90. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006; 6:674–687.91. Hoffmann K, Shibo L, Xiao Z, Longerich T, Büchler MW, Schemmer P. Correlation of gene expression of ATP-binding cassette protein and tyrosine kinase signaling pathway in patients with hepatocellular carcinoma. Anticancer Res. 2011; 31:3883–3890.92. Gollob JA, Wilhelm S, Carter C, Kelley SL. Role of Raf kinase in cancer: therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol. 2006; 33:392–406.93. Feng DY, Zheng H, Tan Y, Cheng RX. Effect of phosphorylation of MAPK and Stat3 and expression of c-fos and c-jun proteins on hepatocarcinogenesis and their clinical significance. World J Gastroenterol. 2001; 7:33–36.94. Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014; 46:1267–1273.95. Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, et al. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011; 54:311–319.96. Freeman AK, Ritt DA, Morrison DK. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol Cell. 2013; 49:751–758.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular assembly of mitogen-activated protein kinase module in ras-transformed NIH3T3 cell line
- Role of Mitogen Activated Protein Kinase and PI 3-kinase on Cell Proliferation in Pancreatic Cancer Cell Lines with K-ras Mutation
- Mitogen-activated Protein Kinase Signaling in Inflammation-related Carcinogenesis
- Anti-proliferative Effect of 15,16-Dihydrotanshinone I Through Cell Cycle Arrest and the Regulation of AMP-activated Protein Kinase/Akt/mTOR and Mitogen-activated Protein Kinase Signaling Pathway in Human Hepatocellular Carcinoma Cells
- Current Status of Molecular Targeted Therapies in Hepatocellular Carcinoma