Lab Med Online.
2020 Apr;10(2):165-168. 10.3343/lmo.2020.10.2.165.
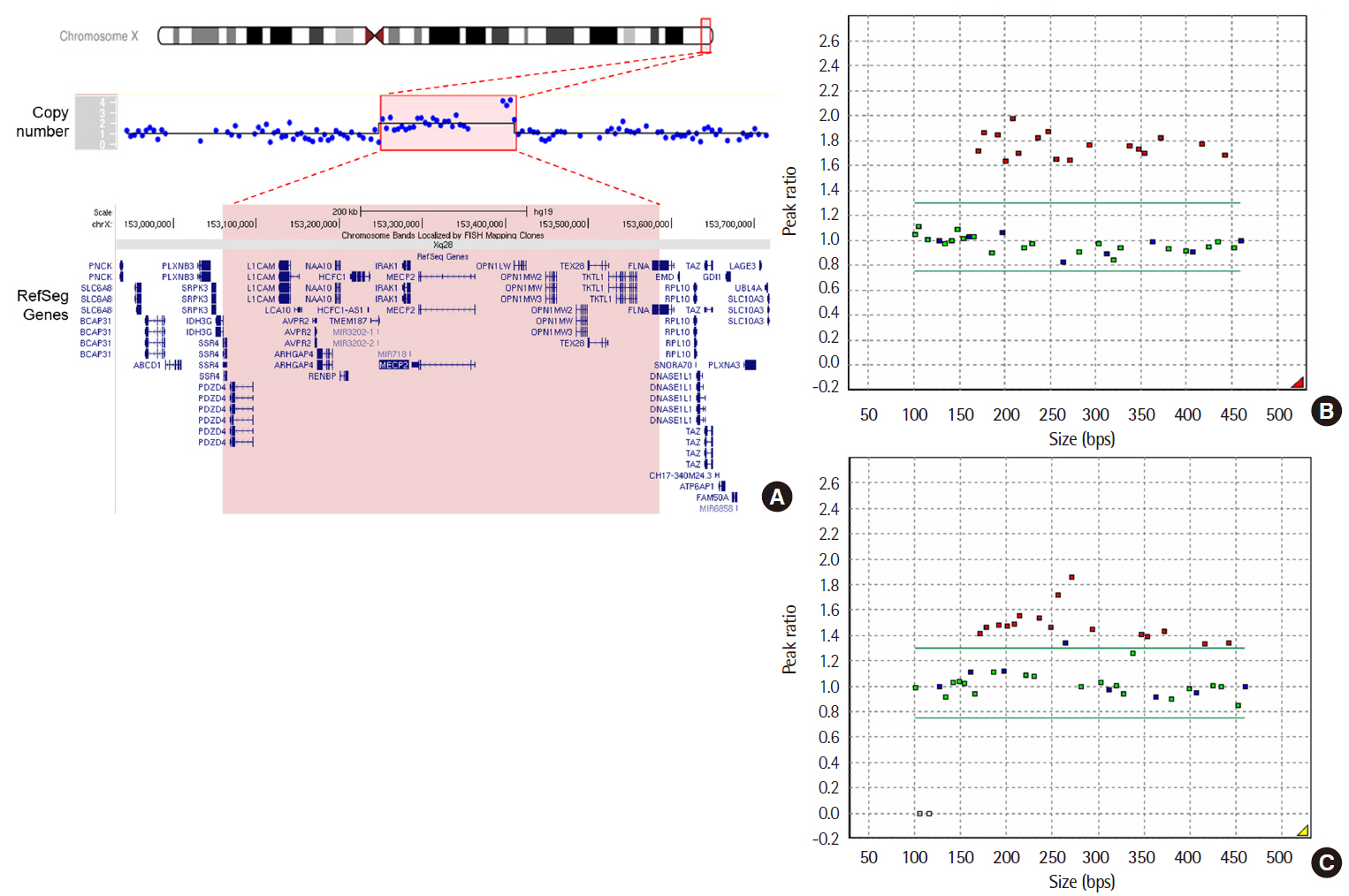
Identification of MECP2 Duplication Using Low-Depth Whole-Genome Sequencing-Based Copy Number Variation Analysis
- Affiliations
-
- 1Department of Laboratory Medicine and Genetics, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea
- 2Department of Pediatrics, Soonchunhyang University Bucheon Hospital, Soonchunhyang University College of Medicine, Bucheon, Korea
- 3Department of Laboratory Medicine, Hanyang University College of Medicine, Guri, Korea
- 4Department of Laboratory Medicine, Hanyang University College of Medicine, Seoul, Korea
- 5GC Genome, Yongin, Korea
- KMID: 2512248
- DOI: http://doi.org/10.3343/lmo.2020.10.2.165
Figure
Reference
-
1. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. 2010; Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 86:749–64. DOI: 10.1016/j.ajhg.2010.04.006. PMID: 20466091. PMCID: PMC2869000.
Article2. Lee JS, Hwang H, Kim SY, Kim KJ, Choi JS, Woo MJ, et al. 2018; Chromosomal microarray with clinical diagnostic utility in children with developmental delay or intellectual disability. Ann Lab Med. 38:473–80. DOI: 10.3343/alm.2018.38.5.473. PMID: 29797819. PMCID: PMC5973923.
Article3. Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. 2013; Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 14:S1. DOI: 10.1186/1471-2105-14-S11-S1. PMID: 24564169. PMCID: PMC3846878.
Article4. Martin-Rodriguez S, Calvo-Ferrer A, Ortega-Unanue N, Samaniego-Jimenez L, Sanz-Izquierdo MP, Bernardo-Gonzalez I. 2019; Two novel variants in the ATM gene causing ataxia-telangiectasia, including a duplication of 90 kb: Utility of targeted next-generation sequencing in de-tection of copy number variation. Ann Hum Genet. 83:266–73. DOI: 10.1111/ahg.12312. PMID: 30888062.5. Jiao Q, Sun H, Zhang H, Wang R, Li S, Sun D, et al. 2019; The combination of whole-exome sequencing and copy number variation sequencing enables the diagnosis of rare neurological disorders. Clin Genet. 96:140–50. DOI: 10.1111/cge.13548. PMID: 30945278.6. Berberich AJ, Huot C, Cao H, Mclntyre AD, Robinson JF, Wang J, et al. 2019; Copy number variation in GCK in patients with maturity-onset diabetes of the young. J Clin Endocrinol Metab. 104:3428–36. DOI: 10.1210/jc.2018-02574. PMID: 30912798. PMCID: PMC6594302.
Article7. Dong Z, Xie W, Chen H, Xu J, Wang H, Li Y, et al. 2017; Copy-number variants detection by low-pass whole-genome sequencing. Curr Protoc Hum Genet. 94:8. DOI: 10.1002/cphg.43. PMID: 28696555.
Article8. Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, et al. 2016; Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med. 18:940–8. DOI: 10.1038/gim.2015.199. PMID: 26820068.
Article9. Venkatraman ES and Olshen AB. 2007; A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 23:657–63. DOI: 10.1093/bioinformatics/btl646. PMID: 17234643.10. Ramocki MB, Tavyev YJ, Peters SU. 2010; The MECP2 duplication syndrome. Am J Med Genet A. 152A:1079–88. DOI: 10.1002/ajmg.a.33184. PMID: 20425814. PMCID: PMC2861792.11. Gonzales ML and LaSalle JM. 2010; The role of MECP2 in brain development and neurodevelopmental disorders. Curr Psychiatry Rep. 12:127–34. DOI: 10.1007/s11920-010-0097-7. PMID: 20425298. PMCID: PMC2847695.12. Clayton-Smith J, Walters S, Hobson E, Burkitt-Wright E, Smith R, Toutain A, et al. 2009; Xq28 duplication presenting with intestinal and bladder dysfunction and a distinctive facial appearance. Eur J Hum Genet. 17:434–43. DOI: 10.1038/ejhg.2008.192. PMID: 18854860. PMCID: PMC2986219.
Article13. Yon DK, Park JE, Kim SJ, Shim SH, Chae KY. 2017; A sibship with duplication of Xq28 inherited from the mother; genomic characterization and clinical outcomes. BMC Med Genet. 18:30. DOI: 10.1186/s12881-017-0394-7. PMID: 28302064. PMCID: PMC5356410.
Article14. Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, et al. 2014; A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 15:256–78. DOI: 10.1093/bib/bbs086. PMID: 23341494. PMCID: PMC3956068.
Article15. Yin AH, Peng CF, Zhao X, Caughey BA, Yang JX, Liu J, et al. 2015; Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA. Proc Natl Acad Sci U S A. 112:14670–5. DOI: 10.1073/pnas.1518151112. PMID: 26554006. PMCID: PMC4664371.
Article16. Lo KK, Karampetsou E, Boustred C, McKay F, Mason S, Hill M, et al. 2016; Limited clinical utility of non-invasive prenatal testing for subchromosomal abnormalities. Am J Hum Genet. 98:34–44. DOI: 10.1016/j.ajhg.2015.11.016. PMID: 26708752. PMCID: PMC4716686.
Article17. Shevell M, Ashwal S, Donley D, Flint J, Gingold M, Hirtz D, et al. 2003; Practice parameter: evaluation of the child with global developmental delay: report of the quality standards subcommittee of the American Academy of Neurology and the practice committee of the Child Neurology Society. Neurology. 60:367–80. DOI: 10.1212/01.WNL.0000031431.81555.16. PMID: 12578916.
Article18. Flore LA and Milunsky JM. 2012; Updates in the genetic evaluation of the child with global developmental delay or intellectual disability. Semin Pediatr Neurol. 19:173–80. DOI: 10.1016/j.spen.2012.09.004. PMID: 23245550.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Detection of hydin Gene Duplication in Personal Genome Sequence Data
- Web-Based Database and Viewer of East Asian Copy Number Variations
- A comprehensive profile of DNA copy number variations in a Korean population: identification of copy number invariant regions among Koreans
- CGHscape: A Software Framework for the Detection and Visualization of Copy Number Alterations
- Array-based Comparative Genomic Hybridization and Its Application to Cancer Genomes and Human Genetics