Cancer Res Treat.
2021 Jan;53(1):9-24. 10.4143/crt.2020.434.
TM4SF4 and LRRK2 Are Potential Therapeutic Targets in Lung and Breast Cancers through Outlier Analysis
- Affiliations
-
- 1Department of Health Sciences and Technology, SAIHST, Sungkyunkwan University, Seoul, Korea
- 2Laboratory of Cancer Genomics and Molecular Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 3College of Pharmacy, Daegu Catholic University, Daegu, Korea
- 4Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea
- 5College of Pharmacy, Duksung Women's University, Seoul, Korea
- 6Interdisciplinary Program in Bioinformatics, College of Natural Science, Seoul National University, Seoul, Korea
- 7Bio-MAX/N-BIO, Seoul National University, Seoul, Korea
- 8Laboratory of Molecular Pathology and Cancer Genomics, Research Institute of Pharmaceutical Sciences and College of Pharmacy, Seoul National University, Seoul, Korea
- 9Department of Otorhinolaryngology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- 10Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2510643
- DOI: http://doi.org/10.4143/crt.2020.434
Abstract
- Purpose
To find biomarkers for disease, there have been constant attempts to investigate the genes that differ from those in the disease groups. However, the values that lie outside the overall pattern of a distribution, the outliers, are frequently excluded in traditional analytical methods as they are considered to be ‘some sort of problem.’ Such outliers may have a biologic role in the disease group. Thus, this study explored new biomarker using outlier analysis, and verified the suitability of therapeutic potential of two genes (TM4SF4 and LRRK2).
Materials and Methods
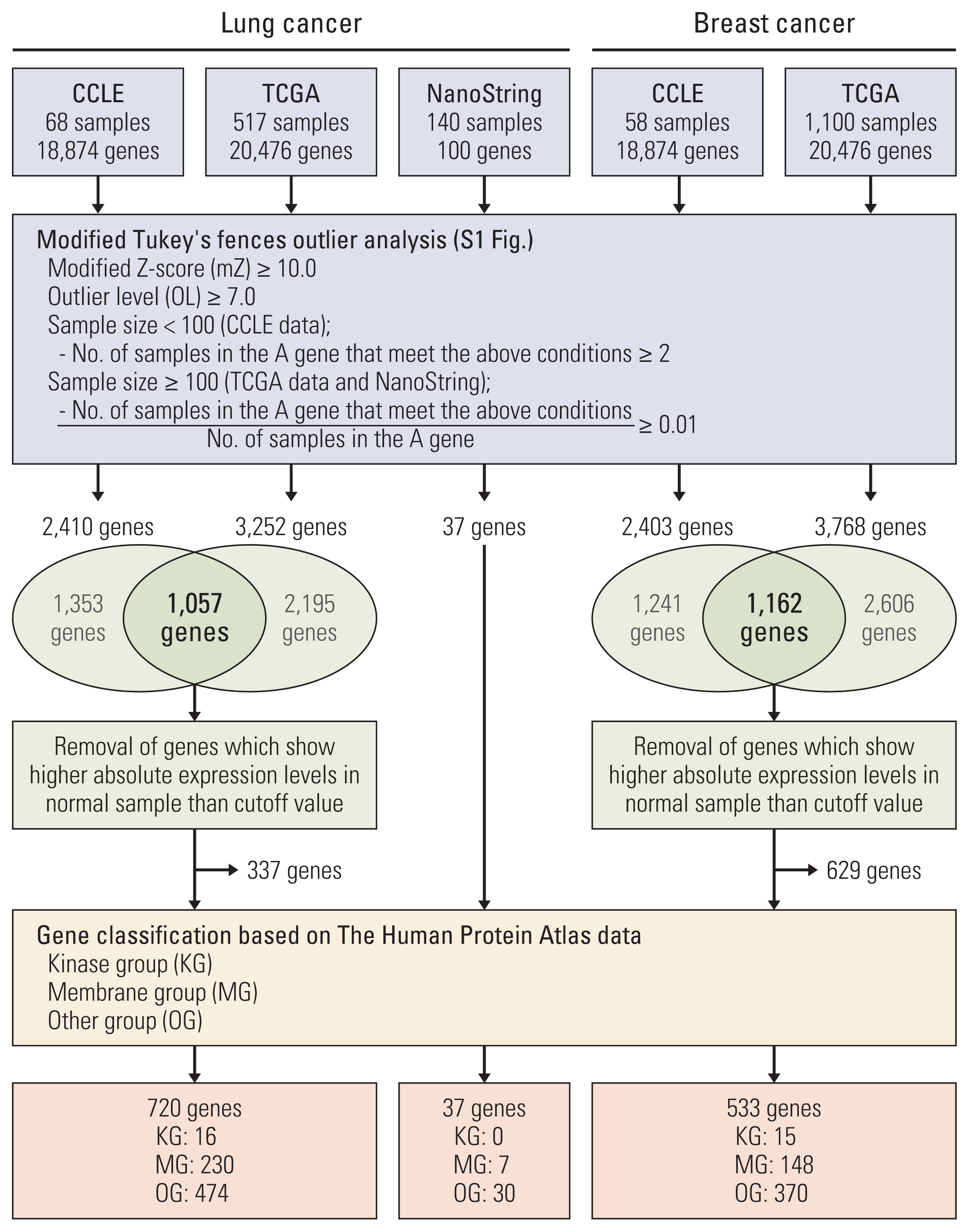
Modified Tukey’s fences outlier analysis was carried out to identify new biomarkers using the public gene expression datasets. And we verified the presence of the selected biomarkers in other clinical samples via customized gene expression panels and tissue microarrays. Moreover, a siRNA-based knockdown test was performed to evaluate the impact of the biomarkers on oncogenic phenotypes.
Results
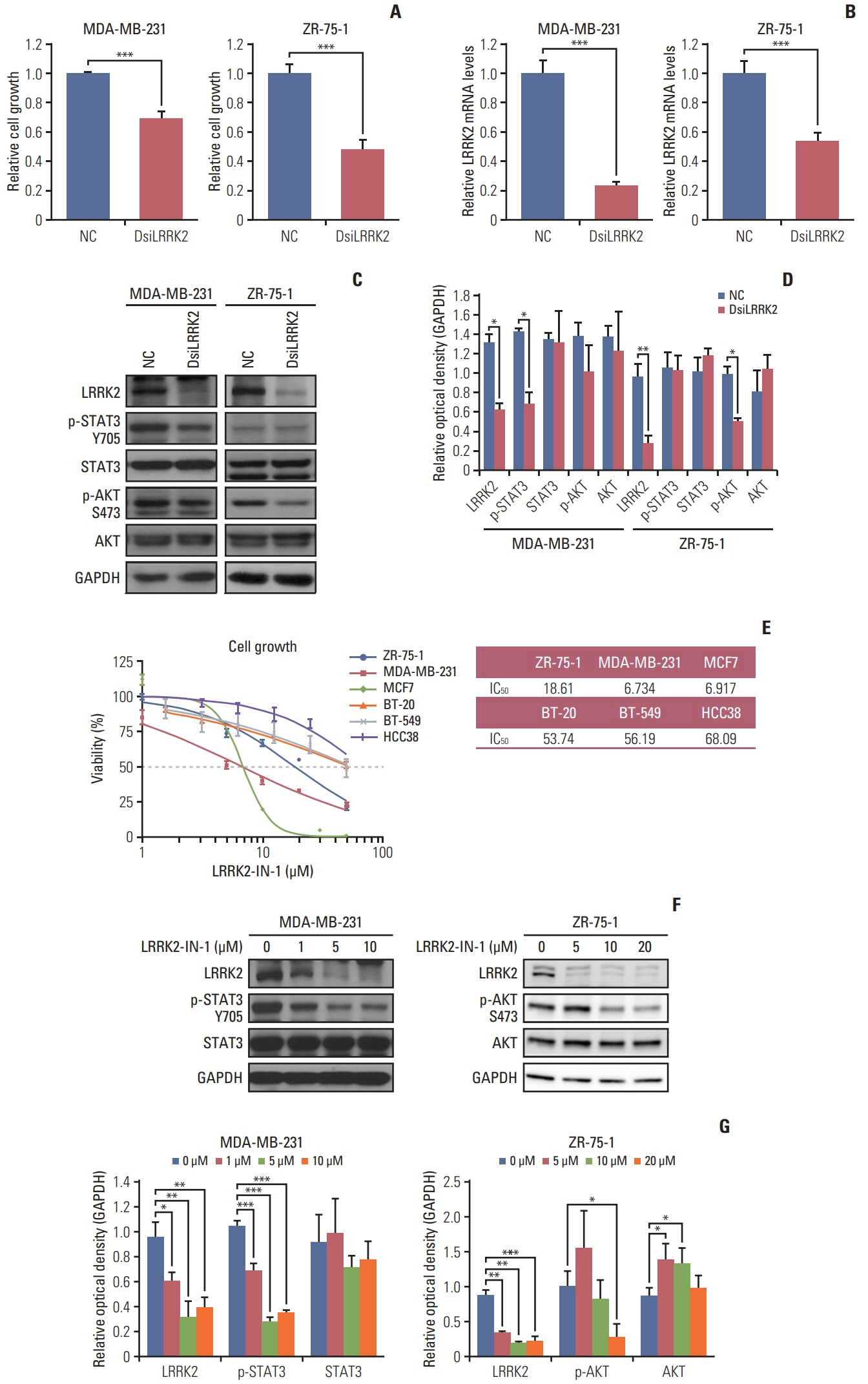
TM4SF4 in lung cancer and LRRK2 in breast cancer were chosen as candidates among the genes derived from the analysis. TM4SF4 and LRRK2 were overexpressed in the small number of samples with lung cancer (4.20%) and breast cancer (2.42%), respectively. Knockdown of TM4SF4 and LRRK2 suppressed the growth of lung and breast cancer cell lines. The LRRK2 overexpressing cell lines were more sensitive to LRRK2-IN-1 than the LRRK2 under-expressing cell lines
Conclusion
Our modified outlier-based analysis method has proved to rescue biomarkers previously missed or unnoticed by traditional analysis showing TM4SF4 and LRRK2 are novel target candidates for lung and breast cancer, respectively.
Keyword
Figure
Reference
-
References
1. Twomey JD, Brahme NN, Zhang B. Drug-biomarker co-development in oncology: 20 years and counting. Drug Resist Updat. 2017; 30:48–62.2. Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005; 310:644–8.
Article3. MacDonald JW, Ghosh D. COPA: cancer outlier profile analysis. Bioinformatics. 2006; 22:2950–1.4. Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015; 6:7002.
Article5. Shim HS, Choi YL, Kim L, Chang S, Kim WS, Roh MS, et al. Molecular testing of lung cancers. J Pathol Transl Med. 2017; 51:242–54.
Article6. Seo S. A review and comparison of methods for detecting outliers in univariate data sets [thesis]. Pittsburgh, PA: University of Pittsburgh;2006.7. Wu A, Wu B, Guo J, Luo W, Wu D, Yang H, et al. Elevated expression of CDK4 in lung cancer. J Transl Med. 2011; 9:38.
Article8. Bronte G, Ulivi P, Verlicchi A, Cravero P, Delmonte A, Crino L. Targeting RET-rearranged non-small-cell lung cancer: future prospects. Lung Cancer (Auckl). 2019; 10:27–36.9. Butti R, Das S, Gunasekaran VP, Yadav AS, Kumar D, Kundu GC. Receptor tyrosine kinases (RTKs) in breast cancer: signaling, therapeutic implications and challenges. Mol Cancer. 2018; 17:34.
Article10. Naidoo K, Wai PT, Maguire SL, Daley F, Haider S, Kriplani D, et al. Evaluation of CDK12 protein expression as a potential novel biomarker for DNA damage response-targeted therapies in breast cancer. Mol Cancer Ther. 2018; 17:306–15.
Article11. Anderson KR, Singer RA, Balderes DA, Hernandez-Lagunas L, Johnson CW, Artinger KB, et al. The L6 domain tetraspanin Tm4sf4 regulates endocrine pancreas differentiation and directed cell migration. Development. 2011; 138:3213–24.
Article12. Choi SI, Kim SY, Lee J, Cho EW, Kim IG. TM4SF4 overexpression in radiation-resistant lung carcinoma cells activates IGF1R via elevation of IGF1. Oncotarget. 2014; 5:9823–37.
Article13. Lim SH, Kim SY, Kim K, Jang H, Ahn S, Kim KM, et al. The implication of FLT3 amplification for FLT targeted therapeutics in solid tumors. Oncotarget. 2017; 8:3237–45.
Article14. Holz MK. The role of S6K1 in ER-positive breast cancer. Cell Cycle. 2012; 11:3159–65.
Article15. Taymans JM, Greggio E. LRRK2 kinase inhibition as a therapeutic strategy for Parkinson’s disease, where do we stand? Curr Neuropharmacol. 2016; 14:214–25.
Article16. Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, et al. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2006; 354:424–5.17. Looyenga BD, Furge KA, Dykema KJ, Koeman J, Swiatek PJ, Giordano TJ, et al. Chromosomal amplification of leucine-rich repeat kinase-2 (LRRK2) is required for oncogenic MET signaling in papillary renal and thyroid carcinomas. Proc Natl Acad Sci U S A. 2011; 108:1439–44.
Article18. Ohta E, Kawakami F, Kubo M, Obata F. LRRK2 directly phosphorylates Akt1 as a possible physiological substrate: impairment of the kinase activity by Parkinson’s disease-associated mutations. FEBS Lett. 2011; 585:2165–70.
Article19. Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol. 2011; 7:203–5.
Article20. Brechtmann F, Mertes C, Matuseviciute A, Yepez VA, Avsec Z, Herzog M, et al. OUTRIDER: a statistical method for detecting aberrantly expressed genes in RNA sequencing data. Am J Hum Genet. 2018; 103:907–17.
Article21. Jeong HM, Kwon MJ, Shin YK. Overexpression of cancer-associated genes via epigenetic derepression mechanisms in gynecologic cancer. Front Oncol. 2014; 4:12.
Article22. Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug resistance in cancer: an overview. Cancers (Basel). 2014; 6:1769–92.
Article23. Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011; 121:3797–803.
Article24. Kim RN, Choi YL, Lee MS, Lira ME, Mao M, Mann D, et al. SEC31A-ALK fusion gene in lung adenocarcinoma. Cancer Res Treat. 2016; 48:398–402.25. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010; 363:1693–703.26. Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J Hematol Oncol. 2017; 10:105.
Article27. Wang L, Feng J, Da L, Li Y, Li Z, Zhao M. Adenovirus-mediated delivery of siRNA targeting TM4SF4 attenuated liver cancer cell growth in vitro and in vivo. Acta Biochim Biophys Sin (Shanghai). 2013; 45:213–9.
Article28. Wallings R, Manzoni C, Bandopadhyay R. Cellular processes associated with LRRK2 function and dysfunction. FEBS J. 2015; 282:2806–26.
Article29. Saunders-Pullman R, Barrett MJ, Stanley KM, Luciano MS, Shanker V, Severt L, et al. LRRK2 G2019S mutations are associated with an increased cancer risk in Parkinson disease. Mov Disord. 2010; 25:2536–41.30. Allegra R, Tunesi S, Cilia R, Pezzoli G, Goldwurm S. LRRK2-G2019S mutation is not associated with an increased cancer risk: a kin-cohort study. Mov Disord. 2014; 29:1325–6.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Pharmacological Inhibition of LRRK2 Exhibits Neuroprotective Activity in Mouse Photothrombotic Stroke Model
- Systems Biology Approaches to Decoding the Genome of Liver Cancer
- Prognostic Value of TZAP Expression in Various Cancers: TCGA Data Analysis
- Rab GTPases as Physiological Substrates of LRRK2 Kinase
- The Role of Long Noncoding RNAs in Antiestrogen Resistance in Breast Cancer: An Overview and Update