Cancer Res Treat.
2020 Oct;52(4):1120-1134. 10.4143/crt.2020.307.
Genetic Alterations in Preinvasive Lung Synchronous Lesions
- Affiliations
-
- 1Division of Statistics, Medical Research Collaborating Center, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Pathology and Translational Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 3Department of Internal Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 4Department of Thoracic and Cardiovascular Surgery, Seoul National University Bundang Hospital, Seongnam, Korea
- KMID: 2507938
- DOI: http://doi.org/10.4143/crt.2020.307
Abstract
- Purpose
Despite advances in treatment, lung cancer remains the leading cause of cancer mortality. This study aimed to characterise genome-wide tumorigenesis events and to understand the hypothesis of the multistep carcinogenesis of lung adenocarcinoma (LUAD)
Materials and Methods
We conducted multiregion whole-exome sequencing of LUAD with synchronous atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ, or minimally invasive adenocarcinoma of 19 samples from three patients to characterize genome-wide tumorigenesis events and validate the hypothesis of the multistep carcinogenesis of LUAD. We identified potential pathogenic mutations preserved in preinvasive lesions and supplemented the finding by allelic variant level from RNA sequencing.
Results
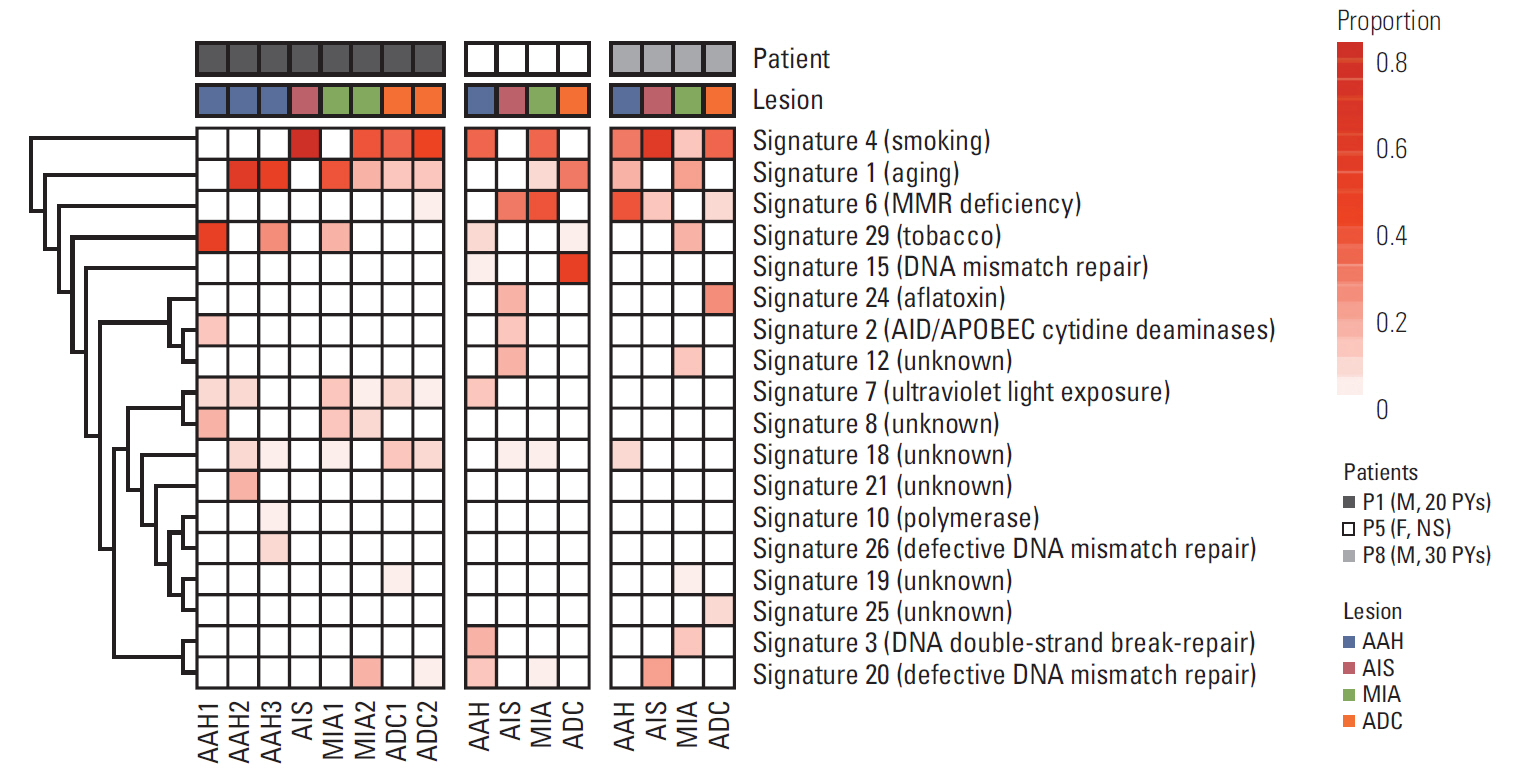
Overall, independent mutational profiles were observed per patient and between patients. Some shared mutations including epidermal growth factor receptor (EGFR , p.L858R) were present across synchronous lesions.
Conclusion
Here, we show that there are driver gene mutations in AAH, and they may exacerbate as a sequence in a histological continuum, supporting the Darwinian evolution model of cancer genome. The intertumoral and intratumoral heterogeneity of synchronous LUAD implies that multi-biomarker strategies might be necessary for appropriate treatment.
Keyword
Figure
Reference
-
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30.
Article2. Pakkala S, Ramalingam SS. Personalized therapy for lung cancer: striking a moving target. JCI Insight. 2018; 3:e120858.
Article3. Choi CM, Kim HC, Jung CY, Cho DG, Jeon JH, Lee JE, et al. Report of the Korean Association of Lung Cancer Registry (KALC-R), 2014. Cancer Res Treat. 2019; 51:1400–10.
Article4. Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014; 346:256–9.
Article5. Travis WD, Nicholson AG. WHO classification of tumours of the lung, pleura, thymus and heart. Lyon: International Agency for Research on Cancer;2015.6. Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci. 2018; 19:1259.
Article7. Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun. 2015; 6:8258.
Article8. Sivakumar S, Lucas FA, McDowell TL, Lang W, Xu L, Fujimoto J, et al. Genomic landscape of atypical adenomatous hyperplasia reveals divergent modes to lung adenocarcinoma. Cancer Res. 2017; 77:6119–30.
Article9. Park E, Ahn S, Kim H, Park SY, Lim J, Kwon HJ, et al. Targeted sequencing analysis of pulmonary adenocarcinoma with multiple synchronous ground-glass/lepidic nodules. J Thorac Oncol. 2018; 13:1776–83.
Article10. Noguchi M. Stepwise progression of pulmonary adenocarcinoma: clinical and molecular implications. Cancer Metastasis Rev. 2010; 29:15–21.11. Tamborero D, Rubio-Perez C, Deu-Pons J, Schroeder MP, Vivancos A, Rovira A, et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018; 10:25.
Article12. Alves JM, Prieto T, Posada D. Multiregional tumor trees are not phylogenies. Trends Cancer. 2017; 3:546–50.
Article13. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018; 173:371–85.14. Rhee JK, Lee S, Park WY, Kim YH, Kim TM. Allelic imbalance of somatic mutations in cancer genomes and transcriptomes. Sci Rep. 2017; 7:1653.
Article15. Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012; 150:1121–34.
Article16. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–50.17. Labbe C, Cabanero M, Korpanty GJ, Tomasini P, Doherty MK, Mascaux C, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer. 2017; 111:23–9.18. Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009; 15:4554–60.
Article19. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–21.20. Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TB, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017; 376:2109–21.21. Valero V 3rd, Pawlik TM, Anders RA. Emerging role of Hpo signaling and YAP in hepatocellular carcinoma. J Hepatocell Carcinoma. 2015; 2:69–78.22. Zheng W, Jiang C, Li R. Integrin and gene network analysis reveals that ITGA5 and ITGB1 are prognostic in non-small-cell lung cancer. Onco Targets Ther. 2016; 9:2317–27.
Article23. Pirinen R, Leinonen T, Bohm J, Johansson R, Ropponen K, Kumpulainen E, et al. Versican in nonsmall cell lung cancer: relation to hyaluronan, clinicopathologic factors, and prognosis. Hum Pathol. 2005; 36:44–50.
Article24. Ma P, Fu Y, Cai MC, Yan Y, Jing Y, Zhang S, et al. Simultaneous evolutionary expansion and constraint of genomic heterogeneity in multifocal lung cancer. Nat Commun. 2017; 8:823.
Article25. He XP, Song FJ, Liu XY, Wang Z, Li XX, Liu FY, et al. The relationship between KRAS gene mutations and HLA class I antigen downregulation in the metastasis of non-small cell lung cancer. J Int Med Res. 2013; 41:1473–83.
Article26. Blakely CM, Watkins TB, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017; 49:1693–704.
Article27. Hakimi AA, Pham CG, Hsieh JJ. A clear picture of renal cell carcinoma. Nat Genet. 2013; 45:849–50.
Article28. Ahn JW, Kim HS, Yoon JK, Jang H, Han SM, Eun S, et al. Identification of somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers. Genome Med. 2014; 6:18.
Article29. Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, et al. Dual chromatin and cytoskeletal remodeling by SETD2. Cell. 2016; 166:950–62.
Article30. Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015; 47:209–16.
Article31. Venkatesan S, Birkbak NJ, Swanton C. Constraints in cancer evolution. Biochem Soc Trans. 2017; 45:1–13.
Article32. Shin HT, Choi YL, Yun JW, Kim NK, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017; 8:1377.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Morphological Features of the Most Advanced Intra-Tumor Component in Multistep Progression
- Usefulness of autofluorescence bronchoscopy for detecting bronchial premalignant lesions
- Synchronous Quadruple Colon Cancer With Two Lesions Previously Obscured by Ischemic Colitis, Plus Bladder Cancer and Thymoma: A Case Report
- Synchronous Primary Lung Cancer with Differrent Squamous cell Carcinoma: One Case Report
- A Case of Primary Synchronous Lung Cancer