J Bacteriol Virol.
2020 Jun;50(2):76-96. 10.4167/jbv.2020.50.2.076.
Lessons Learned from SARS-CoV and MERS-CoV: Preparation for SARS-CoV-2 induced COVID-19
- Affiliations
-
- 1Department of Biotechnology, The Catholic University of Korea, Gyeonggi-do, 14662, Republic of Korea
- KMID: 2504384
- DOI: http://doi.org/10.4167/jbv.2020.50.2.076
Abstract
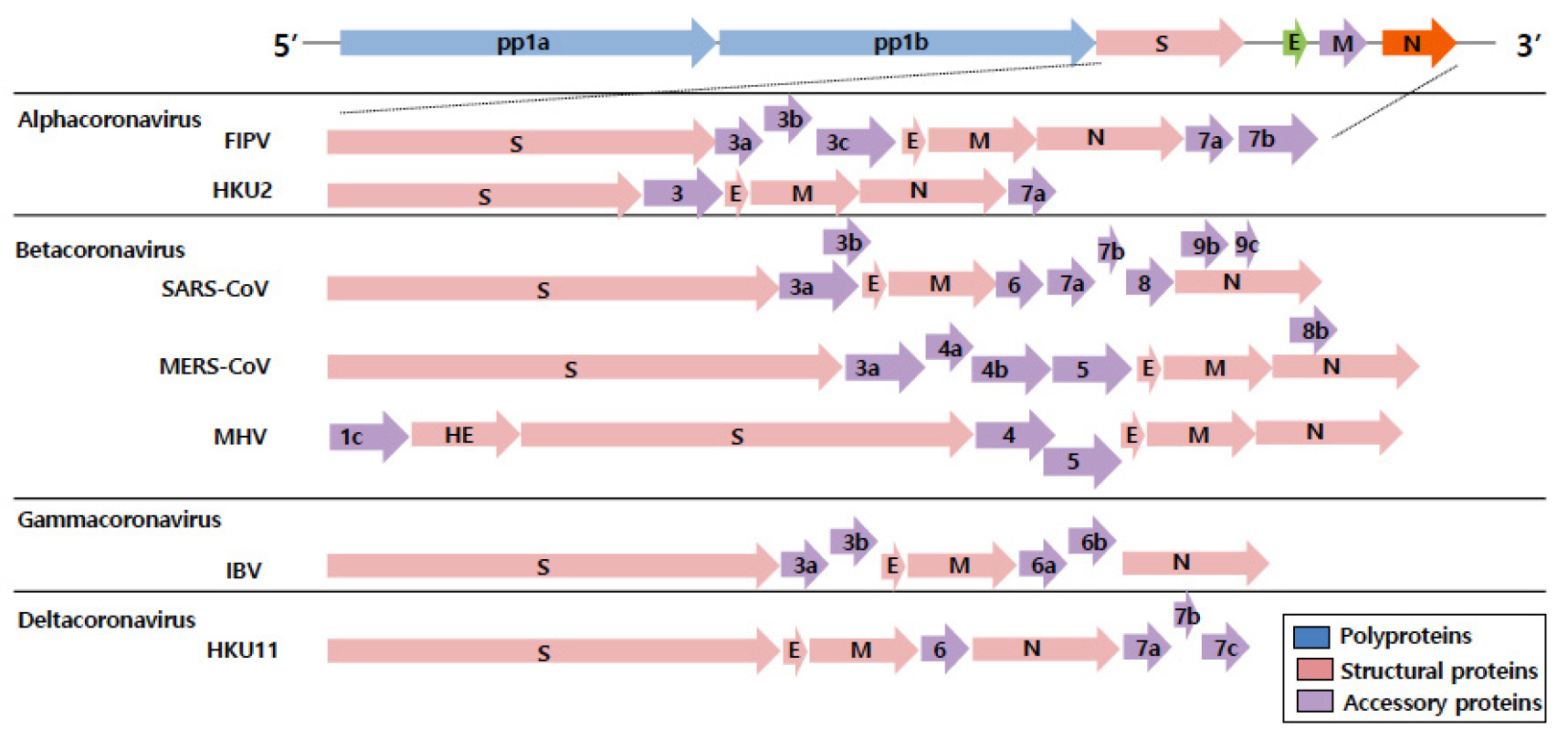
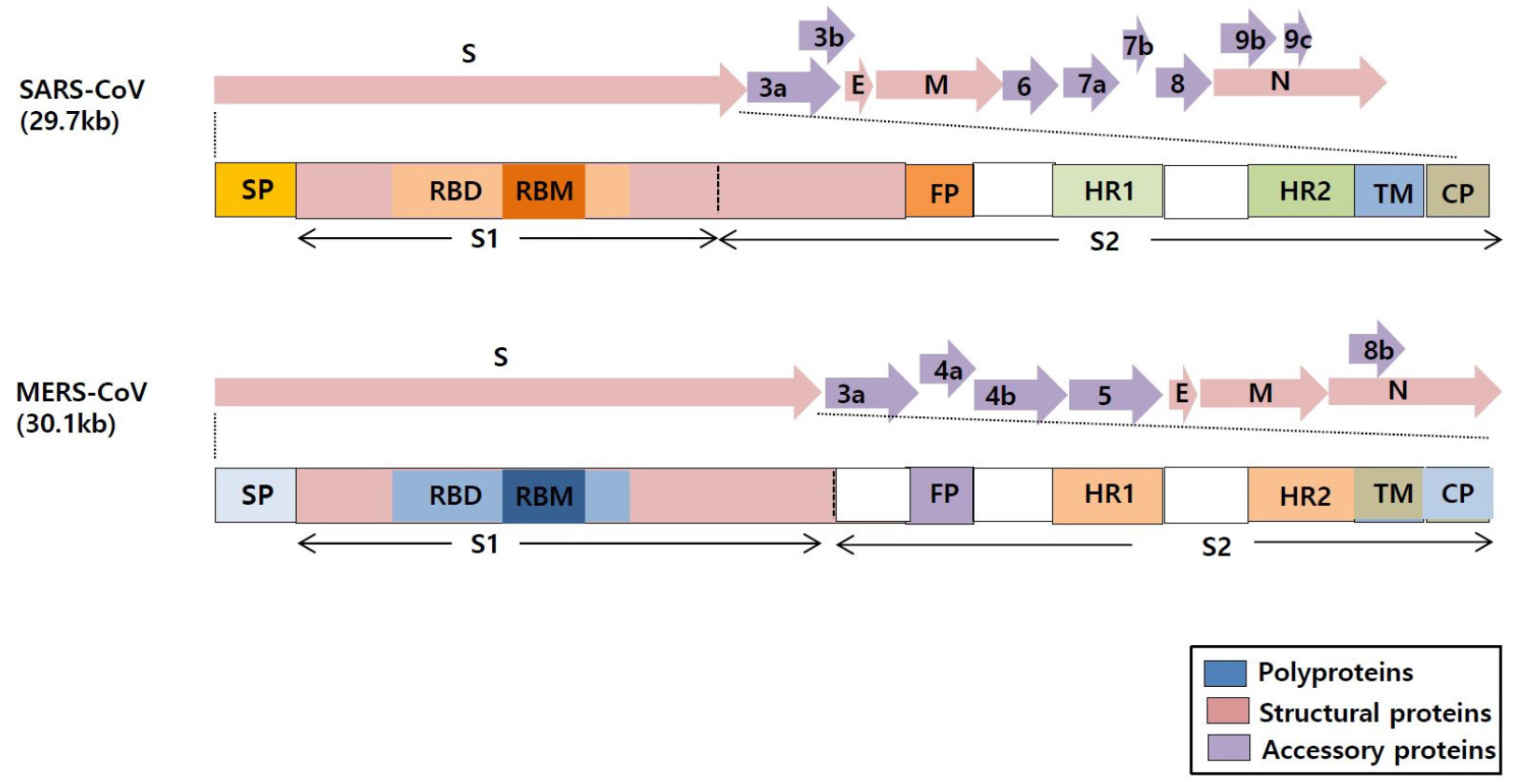
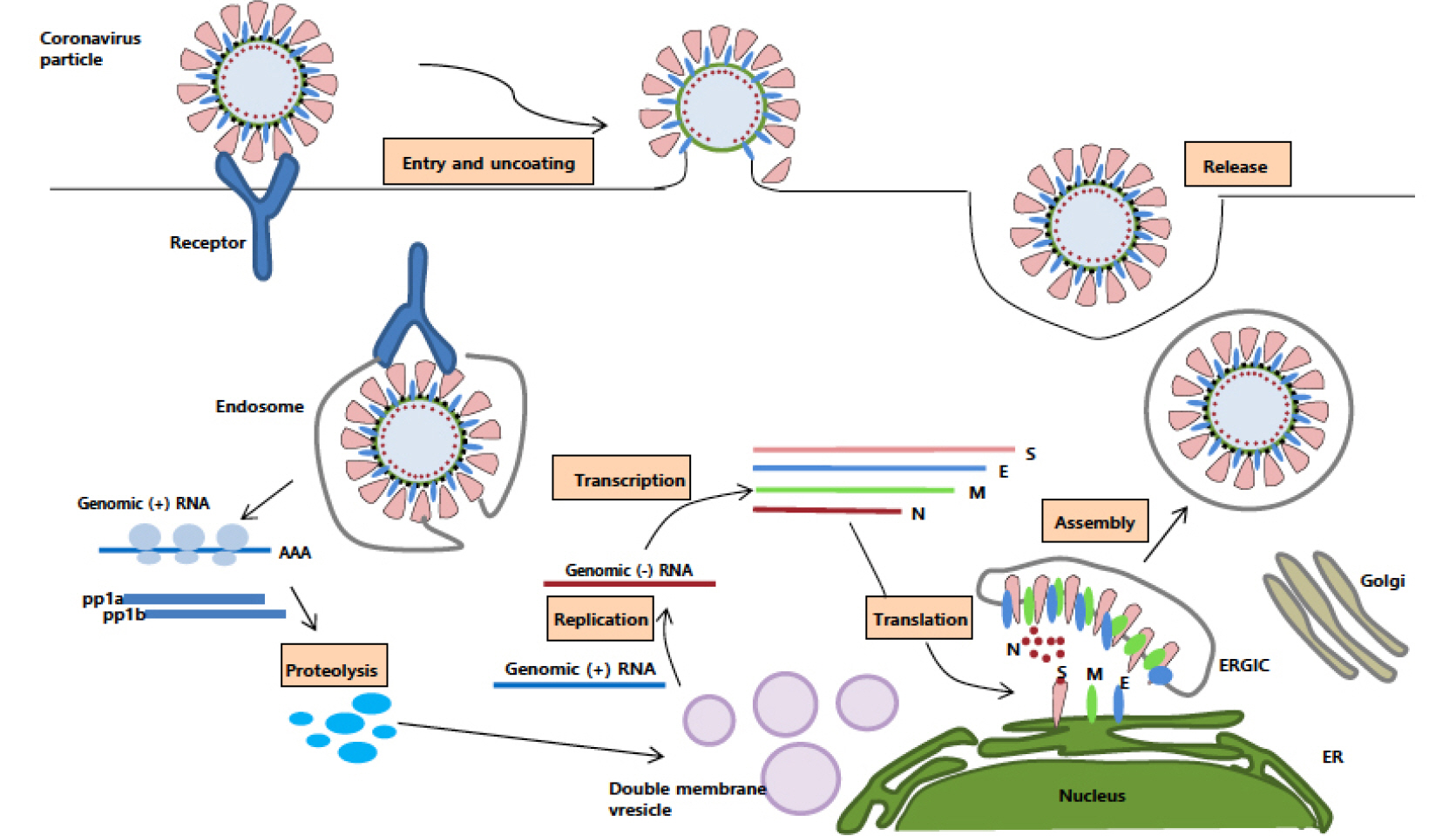
- Coronaviruses (CoVs) are the largest positive-sense RNA viruses with a wide range of natural hosts. To date, seven types of coronaviruses (HCoV-NL63; Human coronavirus NL63, HCoV-229E; Human coronavirus 229E, HCoV-OC43; Human coronavirus OC43, HCoV-HKU1; Human coronavirus HKU1, SARS-CoV; Severe acute respiratory syndrome-related coronavirus, MERS-Co; Middle East respiratory syndrome coronavirus, and SARS-CoV-2; Severe acute respiratory syndrome-related coronavirus) are known to cause disease in humans, and three of the CoVs (SARS-CoV, MERS-CoV, and SARS-CoV-2) cause severe, occasionally fatal, respiratory infections in humans. In November 2002, the case of severe acute respiratory syndrome (SARS), a new respiratory illness caused by SARS-CoV, was first reported in Guangdong Province, China. For the next several months, the SARS outbreak resulted in more than 8,000 cases of infection and 800 deaths. In June 2012, Middle East respiratory syndrome coronavirus (MERS-CoV) was first identified in Saudi Arabia with 2,373 reported viral infections and 823 associated deaths until February 2019. The outbreak of the MERS-CoV pandemic also occurred in South Korea in May 2015. In late December 2019, another novel coronavirus called SARS-CoV-2, genetically linked to SARS-CoV, emerged in Wuhan, Hubei Province of China that has spread worldwide. Outbreaks of coronavirus-infections are occurring frequently in the 21st century; therefore, it seems very likely that another pandemic of coronavirus can emerge anytime in the future. In this review, we outlined the biological characteristics of coronaviruses and summarized the status of vaccine development against SARS-CoV-2, SARS-CoV, and MERS-CoV in preparation for the unpredictable emergence of coronavirus pandemic.
Keyword
Figure
Cited by 1 articles
-
Initial Response of the Korean Society for Laboratory Medicine to the COVID-19 Pandemic
Seung Gyu Yun, Dae-Hyun Ko, Young-Eun Kim, Kyoung Ho Roh, Jaehyeon Lee, Jin-Hee Cho, Misuk Ji, Ki Ho Hong, Hyung-Doo Park, Gye Cheol Kwon
Lab Med Online. 2021;11(4):217-222. doi: 10.47429/lmo.2021.11.4.217.
Reference
-
1. Fung TS, Liu DX. Human Coronavirus: Host-Pathogen Interaction. Annu Rev Microbiol 2019;73:529-57.DOI: 10.1146/annurev-micro-020518-115759. PMID: 31226023.2. Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, Evaluation and Treatment Coronavirus (COVID-19). StatPearls. Treasure Island (FL) 2020.3. Cherry JD, Krogstad P. SARS: the first pandemic of the 21st century. Pediatr Res 2004;56:1-5.DOI: 10.1203/01.PDR.0000129184.87042.FC. PMID: 15152053. PMCID: PMC7086556.4. Hilgenfeld R, Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res 2013;100:286-95.DOI: 10.1016/j.antiviral.2013.08.015. PMID: 24012996. PMCID: PMC7113673.5. https://covid19.who.int.6. CoronaBoard. https://coronaboard.kr/.7. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016;14:523-34.DOI: 10.1038/nrmicro.2016.81. PMID: 27344959. PMCID: PMC7097822.8. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181-92.DOI: 10.1038/s41579-018-0118-9. PMID: 30531947. PMCID: PMC7097006.9. http://www.cdc.go.kr/npt/biz/npp/portal/nppPblctDtaMain.do.10. Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol 2020;92:491-4.DOI: 10.1002/jmv.25709. PMID: 32056249. PMCID: PMC7166760.11. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020;20:363-74.DOI: 10.1038/s41577-020-0311-8. PMID: 32346093. PMCID: PMC7187672.12. Yang M. Cell pryoptosis, a potential pathogenic mechanism of 2019-nCoV Infection. 2020.DOI: 10.2139/ssrn.3527420.13. Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez Med 2020;28:174-84.14. Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019;11:59.DOI: 10.3390/v11010059. PMID: 30646565. PMCID: PMC6357155.15. Tai DY. Pharmacologic treatment of SARS: current knowledge and recommendations. Ann Acad Med Singapore 2007;36:438-43.16. Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med 2006;3:e343.DOI: 10.1371/journal.pmed.0030343. PMID: 16968120. PMCID: PMC1564166.17. Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, et al. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 2003;290:3222-8.DOI: 10.1001/jama.290.24.3222. PMID: 14693875.18. Chu CM, Cheng VC, Hung IF, Wong MM, Chan KH, Chan KS, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 2004;59:252-6.DOI: 10.1136/thorax.2003.012658. PMID: 14985565. PMCID: PMC1746980.19. COVID-19 Studies from the World Health Organization Database. https://clinicaltrials.gov/ct2/who_table.20. Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med 2020;382:1787-99.DOI: 10.1056/NEJMoa2001282. PMID: 32187464. PMCID: PMC7121492.21. Link A, Hold G. First Case of Covid-19 in the United States. N Engl J Med 2020;382:e53.DOI: 10.1056/NEJMc2004794.23. Hu TY, Frieman M, Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat Nanotechnol 2020;15:247-9.DOI: 10.1038/s41565-020-0674-9. PMID: 32203437. PMCID: PMC7094976.24. Coronavirus: France hoping unorthodox virologist can save world. https://www.irishtimes.com/news/world/europe/ coronavirus-france-hoping-unorthodox-virologist-can-save-world-1.4210278.25. Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19?. Int J Antimicrob Agents 2020;55:105938.DOI: 10.1016/j.ijantimicag.2020.105938. PMID: 32171740. PMCID: PMC7118659.26. Japanese flu drug 'clearly effective' in treating coronavirus, says China. https://www.theguardian.com/world/2020/ mar/18/japanese-flu-drug-clearly-effective-intreating-coronavirus-says-china.27. WHO launches global megatrial of the four most promising coronavirus treatments. https://www.sciencemag. org/news/2020/03/who-launches-global-megatrial-four-most-promising-coronavirus-treatments.28. Yong CY, Ong HK, Yeap SK, Ho KL, Tan WS. Recent Advances in the Vaccine Development Against Middle East Respiratory Syndrome-Coronavirus. Front Microbiol 2019;10:1781.DOI: 10.3389/fmicb.2019.01781. PMID: 31428074. PMCID: PMC6688523.29. Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis 2020;ciaa325.DOI: 10.1093/cid/ciaa325. PMID: 32215622. PMCID: PMC7184405.30. Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, et al. "Infection and Rapid Transmission of SARS-CoV-2 in Ferrets." Cell Host Microbe 2020;27:704-9. e2.DOI: 10.1016/j.chom.2020.03.023. PMID: 32259477. PMCID: PMC7144857.31. https://www.biorxiv.org/content/10.1101/2020.05.13.093195v1?fbclid=IwAR1JS6GSibQwse63mGf6lFUnbvb65jj4gTJXiokRVAJ5ZHKELe61gQ38Q9k.32. Zhang J, Zeng H, Gu J, Li H, Zheng L, Zou Q. Progress and Prospects on Vaccine Development against SARS-CoV-2. Vaccines (Basel) 2020;8:E153.DOI: 10.3390/vaccines8020153. PMID: 32235387. PMCID: PMC7349596.33. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 2020;176:104742.DOI: 10.1016/j.antiviral.2020.104742. PMID: 32057769. PMCID: PMC7114094.34. Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A. 2017;114:E7348-57.DOI: 10.1073/pnas.1707304114. PMID: 28807998. PMCID: PMC5584442.35. Muthumani K, Falzarano D, Reuschel EL, Tingey C, Flingai S, Villarreal DO, et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med 2015;7:301ra132.DOI: 10.1126/scitranslmed.aac7462. PMID: 26290414. PMCID: PMC4573558.36. Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol 2016;3:237-61.DOI: 10.1146/annurev-virology-110615-042301. PMID: 27578435. PMCID: PMC5457962.37. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020;581:215-20.DOI: 10.1038/s41586-020-2180-5. PMID: 32225176.38. Chen Y, Guo Y, Pan Y, Zhao ZJ. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun 2020;525:135-40.DOI: 10.1016/j.bbrc.2020.02.071. PMID: 32081428. PMCID: PMC7092824.39. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-3.DOI: 10.1126/science.abb2507. PMID: 32075877. PMCID: PMC7164637.40. Jiang S, He Y, Liu S. SARS vaccine development. Emerg Infect Dis 2005;11:1016-20.DOI: 10.3201/1107.050219. PMID: 16022774. PMCID: PMC3371787.41. Krempl C, Schultze B, Laude H, Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol 1997;71:3285-7.DOI: 10.1128/JVI.71.4.3285-3287.1997. PMID: 9060696. PMCID: PMC191465.42. Chen Y, Lu S, Jia H, Deng Y, Zhou J, Huang B, et al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg Microbes Infect 2017;6:e60.DOI: 10.1038/emi.2017.50. PMID: 28655936. PMCID: PMC5520321.43. Wang Y, Tai W, Yang J, Zhao G, Sun S, Tseng CK, et al. Receptor-binding domain of MERS-CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS-CoV infection. Hum Vaccines Immunother 2017;13:1615-24.DOI: 10.1080/21645515.2017.1296994. PMID: 28277821. PMCID: PMC5512770.44. Adney DR, Wang L, van Doremalen N, Shi W, Zhang Y, Kong WP, et al. Efficacy of an Adjuvanted Middle East Respiratory Syndrome Coronavirus Spike Protein Vaccine in Dromedary Camels and Alpacas. Viruses 2019;11:212.DOI: 10.3390/v11030212. PMID: 30832356. PMCID: PMC6466352.45. Alsaadi EAJ, Neuman BW, Jones IM. A Fusion Peptide in the Spike Protein of MERS Coronavirus. Viruses 2019;11:825.DOI: 10.3390/v11090825. PMID: 31491938. PMCID: PMC6784214.46. Leung DTM, Tam FCH, Ma CH, Chan PKS, Cheung JLK, Niu H, et al. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis 2004;190:379-86.DOI: 10.1086/422040. PMID: 15216476. PMCID: PMC7110057.47. Shi J, Zhang J, Li S, Sun J, Teng Y, Wu M, et al. Epitope-Based Vaccine Target Screening against Highly Pathogenic MERS-CoV: An In Silico Approach Applied to Emerging Infectious Diseases. PLoS One 2015;10:e0144475.DOI: 10.1371/journal.pone.0144475. PMID: 26641892. PMCID: PMC4671582.48. Kim TW, Lee JH, Hung CF, Peng S, Roden R, Wang MC, et al. Generation and characterization of DNA vaccines targeting the nucleocapsid protein of severe acute respiratory syndrome coronavirus. J Virol 2004;78:4638-45.DOI: 10.1128/JVI.78.9.4638-4645.2004. PMID: 15078946. PMCID: PMC387705.49. Collisson EW, Pei J, Dzielawa J, Seo SH. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev Comp Immunol 2000;24:187-200.DOI: 10.1016/S0145-305X(99)00072-5.50. Buchholz UJ, Bukreyev A, Yang L, Lamirande EW, Murphy BR, Subbarao K, et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A 2004;101: 9804-9.DOI: 10.1073/pnas.0403492101. PMID: 15210961. PMCID: PMC470755.51. 10.1101/2020.03.29.2004196252. Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol 2011;174:11-22.DOI: 10.1016/j.jsb.2010.11.021. PMID: 21130884. PMCID: PMC4486061.53. Liu J, Sun Y, Qi J, Chu F, Wu H, Gao F, et al. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J Infect Dis 2010;202:1171-80.DOI: 10.1086/656315. PMID: 20831383.54. Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog 2014;10:e1004077.DOI: 10.1371/journal.ppat.1004077. PMID: 24788150. PMCID: PMC4006877.55. Lee NR, Yi CM, Inn KS. Current advances in the development of vaccines and therapeutic agents against MERS-CoV. J Bacteriol Virol 2015;45:382-8.DOI: 10.4167/jbv.2015.45.4.382.56. Weingartl H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol 2004;78: 12672-6.DOI: 10.1128/JVI.78.22.12672-12676.2004. PMID: 15507655. PMCID: PMC525089.57. Guo JP, Petric M, Campbell W, McGeer PL. SARS corona virus peptides recognized by antibodies in the sera of covalescent case. Virology 2004;324:251-6.DOI: 10.1016/j.virol.2004.04.017. PMID: 15207612. PMCID: PMC7125913.58. Xiong S, Wang YF, Zhang MY, Liu XJ, Zhang CH, Liu SS, et al. Immunogenicity of SARS inactivated vaccine in BALB/c mice. Immunol Lett 2004;95:139-43.DOI: 10.1016/j.imlet.2004.06.014. PMID: 15388253. PMCID: PMC7112924.59. Roper RL, Rehm KE. SARS vaccines: where are we? Expert Rev Vaccines 2009;8:887-98.DOI: 10.1586/erv.09.43. PMID: 19538115. PMCID: PMC7105754.60. See RH, Zakhartchouk AN, Petric M, Lawrence DJ, Mok CP, Hogan RJ, et al. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J Gen Virol 2006;87:641-50.DOI: 10.1099/vir.0.81579-0. PMID: 16476986.61. Darnell ME, Plant EP, Watanabe H, Byrum R, St Claire M, Ward JM, et al. Severe acute respiratory syndrome coronavirus infection in vaccinated ferrets. J Infect Dis 2007;196:1329-38.DOI: 10.1086/522431. PMID: 17922397. PMCID: PMC7110120.62. See RH, Petric M, Lawrence DJ, Mok CPY, Rowe T, Zitzow LA, et al. Severe acute respiratory syndrome vaccine efficacy in ferrets: whole killed virus and adenovirus-vectored vaccines. J Gen Virol 2008;89:2136-46.DOI: 10.1099/vir.0.2008/001891-0. PMID: 18753223.63. Zhou J, Wang W, Zhong Q, Hou W, Yang Z, Xiao SY, et al. Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine 2005;23:3202-9.DOI: 10.1016/j.vaccine.2004.11.075. PMID: 15837221. PMCID: PMC7115379.64. Vetter V, Denizer G, Friedland LR, Krishnan J, Shapiro M. Understanding modern-day vaccines: what you need to know. Ann Med 2018;50:110-20.DOI: 10.1080/07853890.2017.1407035. PMID: 29172780.65. Jung SY, Kang KW, Lee EY, Seo DW, Kim HL, Kim H, et al. Heterologous prime-boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine 2018;36:3468-76.DOI: 10.1016/j.vaccine.2018.04.082. PMID: 29739720. PMCID: PMC7115429.66. Gao W, Tamin A, Soloff A, D'Aiuto L, Nwanegbo E, Robbins PD, et al. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet 2003;362:1895-6.DOI: 10.1016/S0140-6736(03)14962-8.67. https://www.biorxiv.org/content/10.1101/2020.05.13.093195v1.full.68. Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020: S0140-6736(20)31208-3.DOI: 10.1016/S0140-6736(20)31208-3.69. Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 2004;428:561-4.DOI: 10.1038/nature02463. PMID: 15024391. PMCID: PMC7095382.70. Park HJ, Ko HL, Jung SY, Jo HB,Nam JH. The Characteristics of RNA vaccine; its strengths and weaknesses. J Bacteriol Virol 2016;46:115-27.DOI: 10.4167/jbv.2016.46.3.115.71. Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov 2018;17:261-79.DOI: 10.1038/nrd.2017.243. PMID: 29326426. PMCID: PMC5906799.72. Coleman CM, Liu YV, Mu H, Taylor JK, Massare M, Flyer DC, et al. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 2014;32:3169-74.DOI: 10.1016/j.vaccine.2014.04.016. PMID: 24736006. PMCID: PMC4058772.73. Du L, Tai W, Zhou Y, Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev Vaccines 2016;15:1123-34.DOI: 10.1586/14760584.2016.1167603. PMID: 26985862. PMCID: PMC5097835.74. Park H J, Bang EK, Hong JJ, Lee SM, Ko HL, Kwak HW. Nanoformulated Single-Stranded RNA-Based Adjuvant with a Coordinative Amphiphile as an Effective Stabilizer: Inducing Humoral Immune Response by Activation of Antigen-Presenting Cells. Angew Chem Int Ed Engl 2020.DOI: 10.1002/anie.202002979. PMID: 32239636.75. Pang J, Wang MX, Ang IYH, Tan SHX, Lewis RF, Chen JI, et al. Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review. J Clin Med 2020;9:623.DOI: 10.3390/jcm9030623. PMID: 32110875. PMCID: PMC7141113.76. COVID-19 vaccine development pipeline. https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/?fbclid=IwAR2VZ ctyvZI8Xd-a_PAUOPDqtk80q5POzlcH40J8TUxYJ0hmxoW5qN6IU5I.77. Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, et al. The COVID-19 vaccine development landscape. Nat Rev Drug Discov 2020;19:305-6.DOI: 10.1038/d41573-020-00073-5. PMID: 32273591.78. Tu Y F, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int J Mol Sci 2020;21;2657.DOI: 10.3390/ijms21072657. PMID: 32290293. PMCID: PMC7177898.79. Moderna Announces Positive Interim Phase 1 Data for its mRNA Vaccine (mRNA-1273) Against Novel Coronavirus. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-positive-interim-phase-1-data-its-mrna-vaccine\.80. INOVIO Expands Manufacturing of COVID-19 DNA Vaccine INO-4800 With New Funding from CEPI. https:// www.prnewswire.com/news-releases/inovio-expands-manufacturing-of-covid-19-dna-vaccine-ino-4800-with-new-funding-from-cepi-301049889.html.81. Corey L, Mascola JR, Fauci AS, Collins FS. A strategic approach to COVID-19 vaccine R&D. Science 2020;368: 948-950.DOI: 10.1126/science.abc5312. PMID: 32393526.82. Phase I Clinical Trial of a COVID-19 Vaccine in 18-60 Healthy Adults (CTCOVID-19). https://clinicaltrials.gov/ct2/show/ NCT04313127.83. Oxford COVID-19 vaccine to begin phase II/III human trials. http://www.ox.ac.uk/news/2020-05-22-oxford-covid- 19-vaccine-begin-phase-iiiii-human-trials.84. http://www.biospectator.com/view/news_view.php?varAtcId=9624.85. Coronavirus vaccine: Pharma giants GSK and Sanofi team up to find COVID-19 solution. https://www.euronews. com/2020/04/14/coronavirus-vaccine-pharma-giants-gsk-and-sanofi-team-up-to-find-covid-19-solution.86. https://www.medigatenews.com/news/1100597547.87. Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 2012;7:e35421.DOI: 10.1371/journal.pone.0035421. PMID: 22536382. PMCID: PMC3335060.88. Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, Ferris M, et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 2011;85:12201-15.DOI: 10.1128/JVI.06048-11. PMID: 21937658. PMCID: PMC3209347.89. Lurie N, Saville M, Hatchett R, Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N Engl J Med 2020; 382:1969-73.DOI: 10.1056/NEJMp2005630. PMID: 32227757.90. Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 2020; 20:339-41.DOI: 10.1038/s41577-020-0321-6. PMID: 32317716. PMCID: PMC7187142.91. Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 2019;4:e123158.DOI: 10.1172/jci.insight.123158. PMID: 30830861. PMCID: PMC6478436.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Epidemiology, Virology, and Clinical Features of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2; Coronavirus Disease-19)
- Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19)
- COVID-19 and Breastfeeding
- Newly Emerging Human Coronaviruses: Animal Models and Vaccine Research for SARS, MERS, and COVID-19
- Changes in SARS-CoV-2 antibody titers 6 months after the booster dose of BNT162b2 COVID-19 vaccine among health care workers