J Korean Neurosurg Soc.
2020 May;63(3):386-396. 10.3340/jkns.2019.0084.
Spine Computed Tomography to Magnetic Resonance Image Synthesis Using Generative Adversarial Networks : A Preliminary Study
- Affiliations
-
- 1Department of Neurosurgery, Pusan National University Hospital, Busan, Korea
- 2Department of Radiology, Pusan National University Hospital, Busan, Korea
- 3Team Elysium Inc., Seoul, Korea
- 4School of Information and Communication Engineering, Inha University, Incheon, Korea
- KMID: 2501727
- DOI: http://doi.org/10.3340/jkns.2019.0084
Abstract
Objective
: To generate synthetic spine magnetic resonance (MR) images from spine computed tomography (CT) using generative adversarial networks (GANs), as well as to determine the similarities between synthesized and real MR images.
Methods
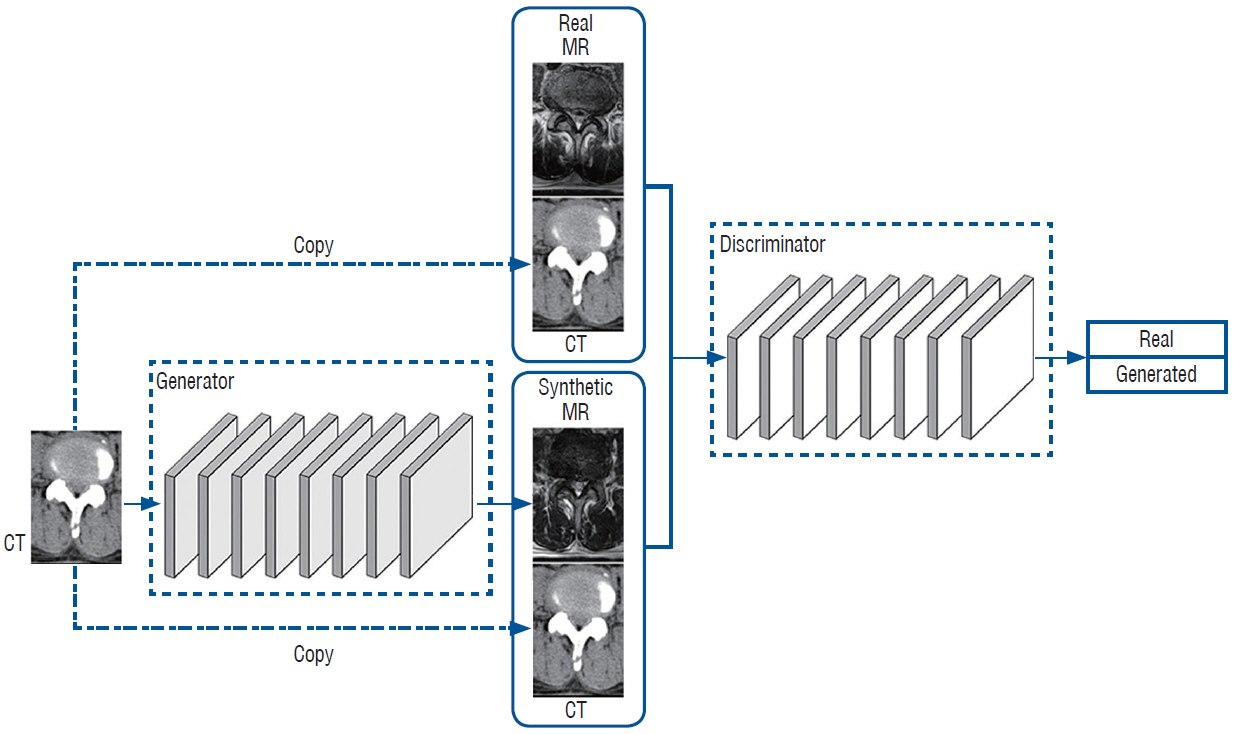
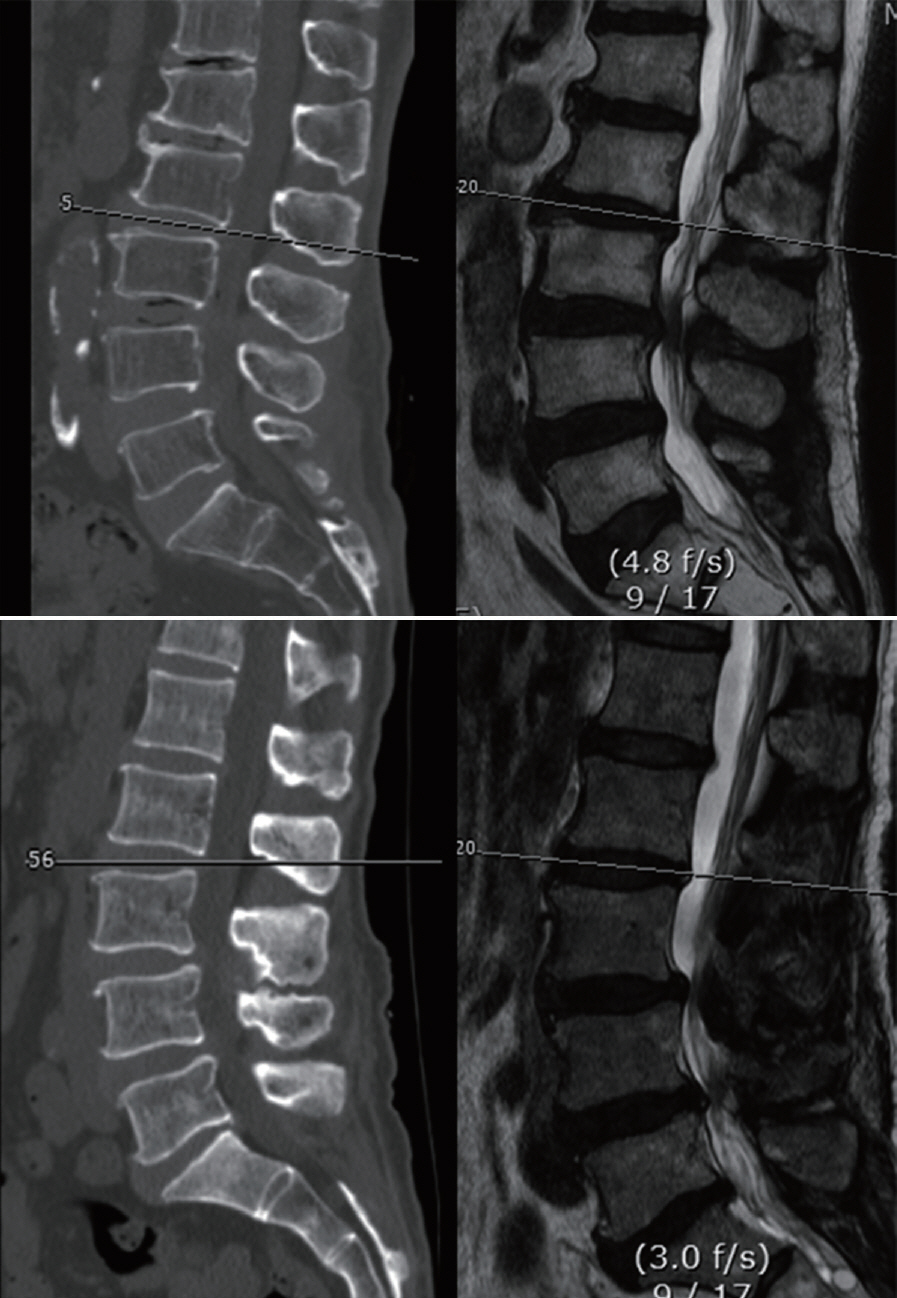
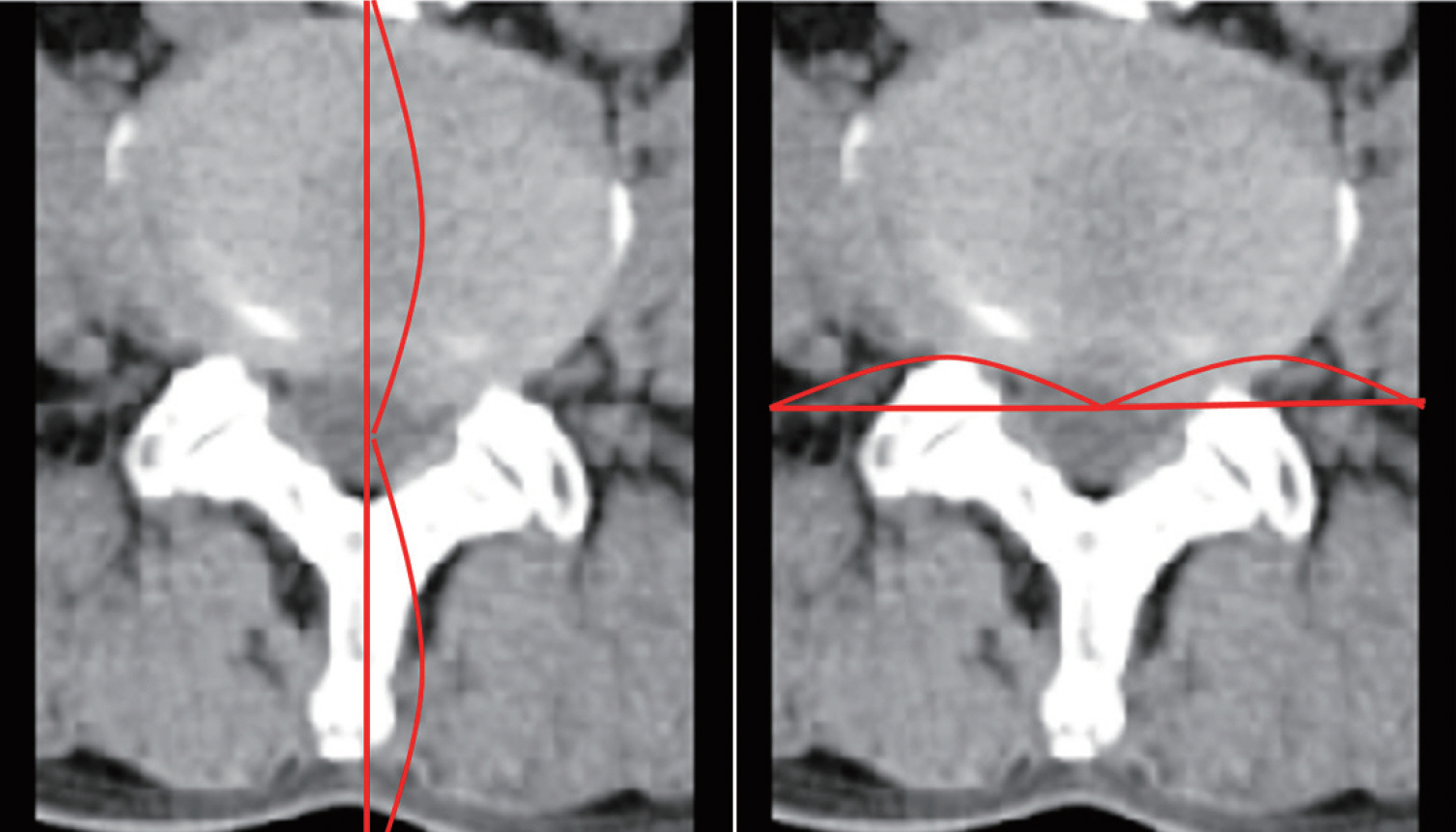
: GANs were trained to transform spine CT image slices into spine magnetic resonance T2 weighted (MRT2) axial image slices by combining adversarial loss and voxel-wise loss. Experiments were performed using 280 pairs of lumbar spine CT scans and MRT2 images. The MRT2 images were then synthesized from 15 other spine CT scans. To evaluate whether the synthetic MR images were realistic, two radiologists, two spine surgeons, and two residents blindly classified the real and synthetic MRT2 images. Two experienced radiologists then evaluated the similarities between subdivisions of the real and synthetic MRT2 images. Quantitative analysis of the synthetic MRT2 images was performed using the mean absolute error (MAE) and peak signal-to-noise ratio (PSNR).
Results
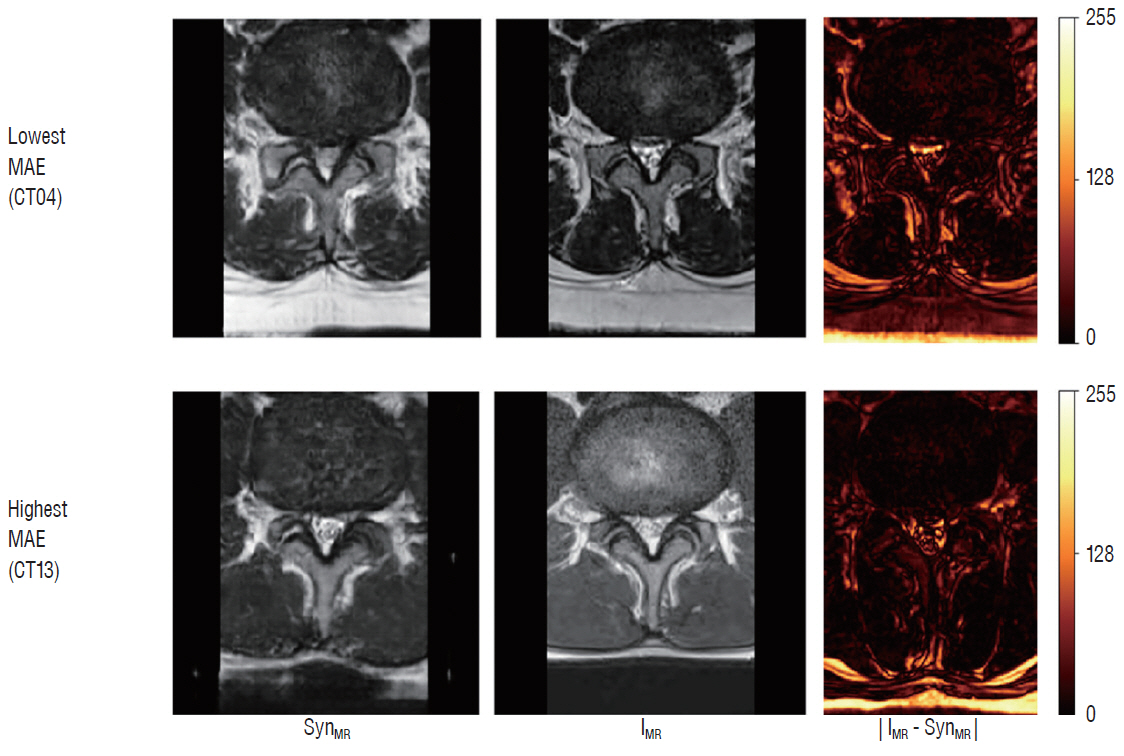
: The mean overall similarity of the synthetic MRT2 images evaluated by radiologists was 80.2%. In the blind classification of the real MRT2 images, the failure rate ranged from 0% to 40%. The MAE value of each image ranged from 13.75 to 34.24 pixels (mean, 21.19 pixels), and the PSNR of each image ranged from 61.96 to 68.16 dB (mean, 64.92 dB).
Conclusion
: This was the first study to apply GANs to synthesize spine MR images from CT images. Despite the small dataset of 280 pairs, the synthetic MR images were relatively well implemented. Synthesis of medical images using GANs is a new paradigm of artificial intelligence application in medical imaging. We expect that synthesis of MR images from spine CT images using GANs will improve the diagnostic usefulness of CT. To better inform the clinical applications of this technique, further studies are needed involving a large dataset, a variety of pathologies, and other MR sequence of the lumbar spine.
Figure
Cited by 1 articles
-
Neurosurgical Management of Cerebrospinal Tumors in the Era of Artificial Intelligence : A Scoping Review
Kuchalambal Agadi, Asimina Dominari, Sameer Saleem Tebha, Asma Mohammadi, Samina Zahid
J Korean Neurosurg Soc. 2023;66(6):632-641. doi: 10.3340/jkns.2021.0213.
Reference
-
References
1. Beechar VB, Zinn PO, Heck KA, Fuller GN, Han I, Patel AJ, et al. Spinal epidermoid tumors: case report and review of the literature. Neurospine. 15:117–122. 2018.
Article2. Bi L, Kim J, Kumar A, Feng D, Fulham M. Synthesis of Positron Emission Tomography (PET) Images via Multi-channel Generative Adversarial Networks (GANs). In : Cardoso MJ, Arbel T, editors. Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment. Cham: Springer;2017. p. 43–51.3. Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: an overview. IEEE Signal Processing Magazine. 35:53–65. 2018.
Article4. Feng R, Badgeley M, Mocco J, Oermann EK. Deep learning guided stroke management: a review of clinical applications. J Neurointerv Surg. 10:358–362. 2018.
Article5. Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 27:2672–2680. 2014.6. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In : Proc IEEE Int Conf Comput Vis; p. 770–778. 2016.
Article7. Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 21:653–660. 2018.
Article8. Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. In : Proc IEEE Int Conf Comput Vis; p. 1125–1134. 2017.
Article9. Jin CB, Kim H, Jung W, Joo S, Park E, Saem AY, et al. Deep CT to MR synthesis using paired and unpaired data. Sensors (Basel). 19:2019. 2019.
Article10. Johnson J, Alahi A, Fei-Fei L. Perceptual Losses for Real-Time Style Transfer and Super-Resolution. In : Leibe B, Matas J, Sebe N, Welling M, editors. Computer Vision - ECCV 2016. Cham: Springer;2016. p. 694–711.11. Kingma DP, Ba J. Adam: a method for stochastic optimization. Available at : https://arxiv.org/abs/1412.6980.12. Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, et al. Medical image synthesis with context-aware generative adversarial networks. Med Image Comput Comput Assist Interv. 10435:417–425. 2017.
Article13. Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, et al. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. 88:581–586. 2017.
Article14. Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA. Context encoders: feature learning by inpainting. In : Proc IEEE Int Conf Comput Vis; p. 2536–2544. 2016.
Article15. Ulyanov D, Vedaldi A, Lempitsky V. Instance normalization: the missing ingredient for fast stylization. Available at : https://arxiv.org/abs/1607.08022.16. Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I. Deep MR to CT synthesis using unpaired data. Available at : https://arxiv.org/abs/1708.01155.
Article17. Xu B, Wang N, Chen T, Li M. Empirical evaluation of rectified activations in convolutional network. Available at : https://arxiv.org/abs/1505.00853.18. Zhao R, Zhao JJ, Dai F, Zhao FQ. A new image secret sharing scheme to identify cheaters. Computer Standards & Interfaces. 31:252–257. 2009.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Uncover This Tech Term: Generative Adversarial Networks
- Image-Based Generative Artificial Intelligence in Radiology: Comprehensive Updates
- Synthesis of T2-weighted images from proton density images using a generative adversarial network in a temporomandibular joint magnetic resonance imaging protocol
- Image Study of the Lumbar Spine
- An Overview of Deep Learning Algorithms and Their Applications in Neuropsychiatry