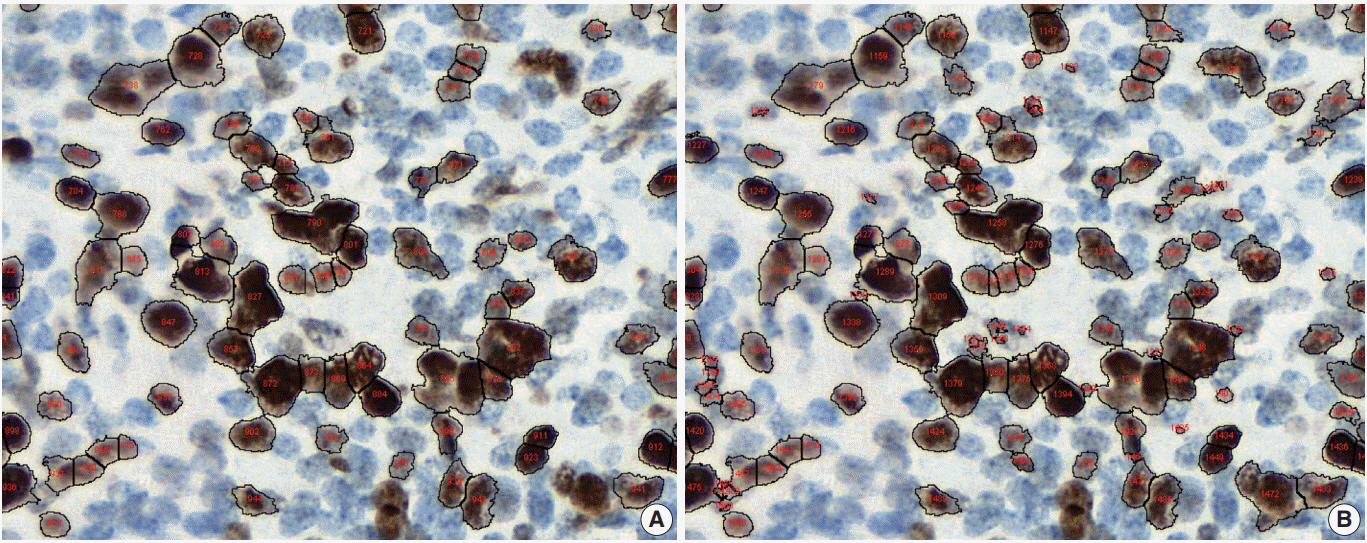
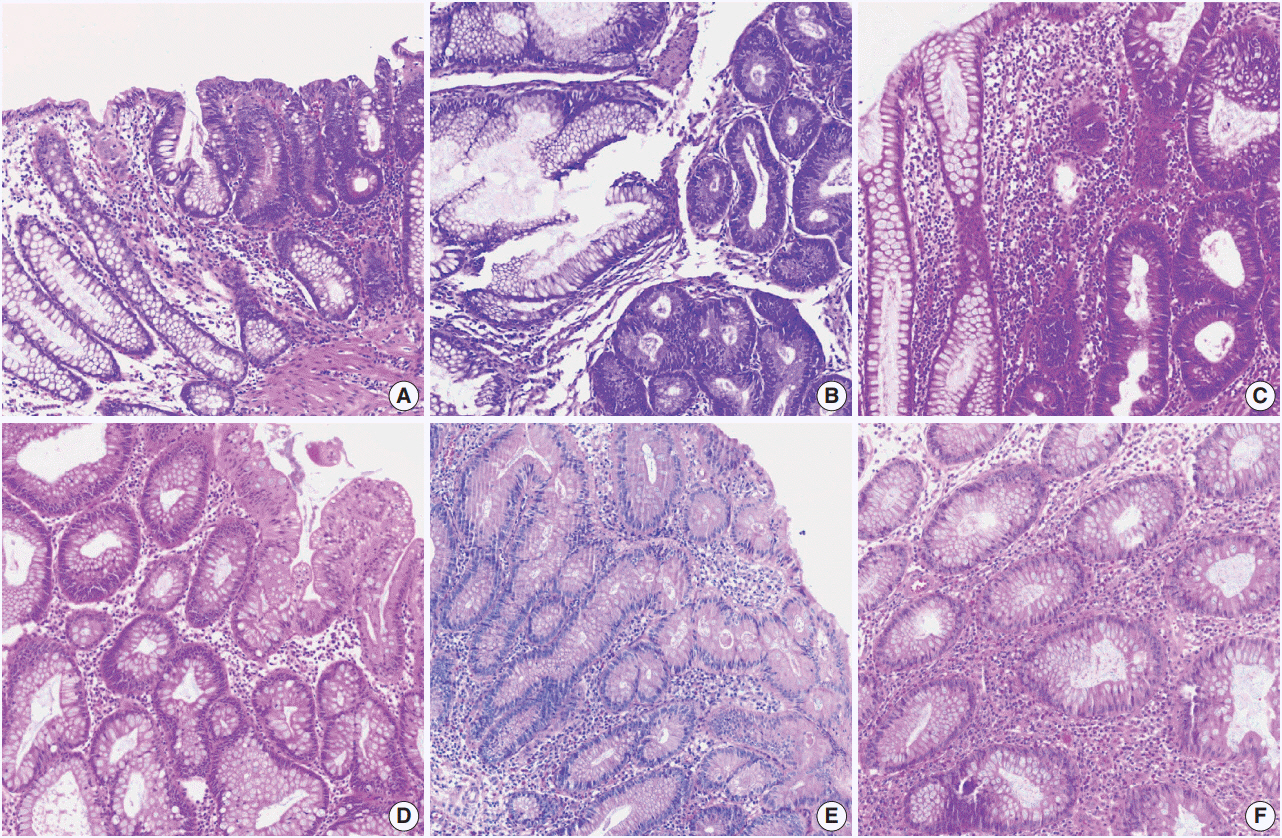
Introduction to digital pathology and computer-aided pathology
- Affiliations
-
- 1Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 3Deep Bio Inc., Seoul, Korea
- 4Department of Urology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 5Catholic Cancer Research Institute, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 6Department of Biomedicine & Health Sciences, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2501602
- DOI: http://doi.org/10.4132/jptm.2019.12.31
Abstract
- Digital pathology (DP) is no longer an unfamiliar term for pathologists, but it is still difficult for many pathologists to understand the engineering and mathematics concepts involved in DP. Computer-aided pathology (CAP) aids pathologists in diagnosis. However, some consider CAP a threat to the existence of pathologists and are skeptical of its clinical utility. Implementation of DP is very burdensome for pathologists because technical factors, impact on workflow, and information technology infrastructure must be considered. In this paper, various terms related to DP and computer-aided pathologic diagnosis are defined, current applications of DP are discussed, and various issues related to implementation of DP are outlined. The development of computer-aided pathologic diagnostic tools and their limitations are also discussed.
Figure
Cited by 3 articles
-
A machine-learning expert-supporting system for diagnosis prediction of lymphoid neoplasms using a probabilistic decision-tree algorithm and immunohistochemistry profile database
Yosep Chong, Ji Young Lee, Yejin Kim, Jingyun Choi, Hwanjo Yu, Gyeongsin Park, Mee Yon Cho, Nishant Thakur
J Pathol Transl Med. 2020;54(6):462-470. doi: 10.4132/jptm.2020.07.11.Recommendations for pathologic practice using digital pathology: consensus report of the Korean Society of Pathologists
Yosep Chong, Dae Cheol Kim, Chan Kwon Jung, Dong-chul Kim, Sang Yong Song, Hee Jae Joo, Sang-Yeop Yi
J Pathol Transl Med. 2020;54(6):437-452. doi: 10.4132/jptm.2020.08.27.Development of quality assurance program for digital pathology by the Korean Society of Pathologists
Yosep Chong, Jeong Mo Bae, Dong Wook Kang, Gwangil Kim, Hye Seung Han
J Pathol Transl Med. 2022;56(6):370-382. doi: 10.4132/jptm.2022.09.30.
Reference
-
1. Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology: new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019; 16:703–15.2. Abels E, Pantanowitz L, Aeffner F, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol. 2019; 249:286–94.
Article3. he j, baxter sl, xu j, xu j, zhou x, zhang k. the practical implementation of artificial intelligence technologies in medicine. nat med. 2019; 25:30–6.
Article4. Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017; 141:944–59.
Article5. Mukhopadhyay S, Feldman MD, Abels E, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: a multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am J Surg Pathol. 2018; 42:39–52.6. Niazi MK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019; 20:e253–61.
Article7. Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017; 69S:S36–40.
Article8. Simon P. Too big to ignore: the business case for big data. Hoboken: John Wiley & Sons;2013.9. Yao X. Evolving artificial neural networks. Proc IEEE. 1999; 87:1423–47.10. Deng L, Yu D. Deep learning: methods and applications. Found Trends Signal Process. 2014; 7:197–387.
Article11. Collobert R, Weston J. A unified architecture for natural language processing: deep neural networks with multitask learning. In : Proceedings of the 25th International Conference on Machine Learning; 2008 Jul 5-9; Helsinki, Finland. New York: Association for Computing Machinery;2008. 160–7.
Article12. Wang S, Yang DM, Rong R, et al. Artificial intelligence in lung cancer pathology image analysis. Cancers (Basel). 2019; 11:E1673.
Article13. Garcia-Rojo M. International clinical guidelines for the adoption of digital pathology: a review of technical aspects. Pathobiology. 2016; 83:99–109.
Article14. Hanna MG, Reuter VE, Samboy J, et al. Implementation of digital pathology offers clinical and operational increase in efficiency and cost savings. Arch Pathol Lab Med. 2019; 143:1545–55.
Article15. Hanna MG, Pantanowitz L, Evans AJ. Overview of contemporary guidelines in digital pathology: what is available in 2015 and what still needs to be addressed? J Clin Pathol. 2015; 68:499–505.
Article16. Snead DR, Tsang YW, Meskiri A, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016; 68:1063–72.
Article17. Irshad H, Veillard A, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: a review-current status and future potential. IEEE Rev Biomed Eng. 2014; 7:97–114.
Article18. Wang S, Yang DM, Rong R, Zhan X, Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol. 2019; 189:1686–98.
Article19. Gurcan MN, Pan T, Shimada H, Saltz J. Image analysis for neuroblastoma classification: segmentation of cell nuclei. Conf Proc IEEE Eng Med Biol Soc. 2006; 1:4844–7.
Article20. Ballaro B, Florena AM, Franco V, Tegolo D, Tripodo C, Valenti C. An automated image analysis methodology for classifying megakaryocytes in chronic myeloproliferative disorders. Med Image Anal. 2008; 12:703–12.21. Korde VR, Bartels H, Barton J, Ranger-Moore J. Automatic segmentation of cell nuclei in bladder and skin tissue for karyometric analysis. Anal Quant Cytol Histol. 2009; 31:83–9.
Article22. Veta M, van Diest PJ, Kornegoor R, Huisman A, Viergever MA, Pluim JP. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PLoS One. 2013; 8:e70221.23. Ruifrok AC, Katz RL, Johnston DA. Comparison of quantification of histochemical staining by hue-saturation-intensity (HSI) transformation and color-deconvolution. Appl Immunohistochem Mol Morphol. 2003; 11:85–91.
Article24. Flanagan MB, Dabbs DJ, Brufsky AM, Beriwal S, Bhargava R. Histopathologic variables predict Oncotype DX recurrence score. Mod Pathol. 2008; 21:1255–61.
Article25. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010; 134:e48–72.26. Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010; 57:841–52.
Article27. stalhammar g, fuentes martinez n, lippert m, et al. digital image analysis outperforms manual biomarker assessment in breast cancer. mod pathol. 2016; 29:318–29.
Article28. Zarrella ER, Coulter M, Welsh AW, et al. Automated measurement of estrogen receptor in breast cancer: a comparison of fluorescent and chromogenic methods of measurement. Lab Invest. 2016; 96:1016–25.
Article29. Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 2016; 33:170–5.
Article30. Williams BJ, Bottoms D, Treanor D. Future-proofing pathology: the case for clinical adoption of digital pathology. J Clin Pathol. 2017; 70:1010–8.
Article31. Kloppel G, La Rosa S. Ki67 labeling index: assessment and prognostic role in gastroenteropancreatic neuroendocrine neoplasms. Virchows Arch. 2018; 472:341–9.
Article32. Chai SM, Brown IS, Kumarasinghe MP. Gastroenteropancreatic neuroendocrine neoplasms: selected pathology review and molecular updates. Histopathology. 2018; 72:153–67.
Article33. Mok TS, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019; 393:1819–30.34. Steven A, Fisher SA, Robinson BW. Immunotherapy for lung cancer. Respirology. 2016; 21:821–33.
Article35. Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015; 125:3335–7.
Article36. Khan AM, Rajpoot N, Treanor D, Magee D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans Biomed Eng. 2014; 61:1729–38.
Article37. Bejnordi BE, Litjens G, Timofeeva N, et al. Stain specific standardization of whole-slide histopathological images. IEEE Trans Med Imaging. 2016; 35:404–15.38. Vahadane A, Peng T, Sethi A, et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging. 2016; 35:1962–71.
Article39. Zarella MD, Yeoh C, Breen DE, Garcia FU. An alternative reference space for H&E color normalization. PLoS One. 2017; 12:e0174489.40. Miyato T, Kataoka T, Koyama M, Yoshida Y. pectral normalization for generative adversarial networks. Preprint at: https://arxiv.org/abs/1802.05957 (2018).41. Shaban MT, Baur C, Navab N, Albarqouni S. StainGAN: stain style transfer for digital histological images. Preprint at: https://arxiv.org/abs/1804.01601 (2018).
Article42. Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the Digital Pathology Association. J Pathol Inform. 2019; 10:9.
Article43. Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019; 25:1301–9.
Article44. Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016; 35:1313–21.
Article45. Irshad H, Montaser-Kouhsari L, Waltz G, et al. Crowdsourcing image annotation for nucleus detection and segmentation in computational pathology: evaluating experts, automated methods, and the crowd. Pac Symp Biocomput. 2015; 294–305.
Article46. Aeffner F, Wilson K, Bolon B, et al. Commentary: roles for pathologists in a high-throughput image analysis Team. Toxicol Pathol. 2016; 44:825–34.47. Tizhoosh HR, Pantanowitz L. Artificial intelligence and digital pathology: challenges and opportunities. J Pathol Inform. 2018; 9:38.
Article48. Serag A, Ion-Margineanu A, Qureshi H, et al. Translational AI and deep learning in diagnostic pathology. Front Med (Lausanne). 2019; 6:185.
Article49. Chang HY, Jung CK, Woo JI, et al. Artificial intelligence in pathology. J Pathol Transl Med. 2019; 53:1–12.
Article50. Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial intelligence and machine learning in pathology: the present landscape of supervised methods. Acad Pathol. 2019; 6:2374289519873088.
Article51. U.S. Food and Drug Administration. Artificial intelligence and machine learning in software as a medical device. Silverspring: U.S. Food and Drug Administration;2019.52. Molnar C. Interpretable machine learning [Internet]. The Author, 2018 [cited 2019 Nov 2]. Available from: https://christophm.github.io/interpretable-ml-book/.53. Kahng M, Andrews PY, Kalro A, Polo Chau DH. ACTIVIS: visual exploration of industry-scale deep neural network models. IEEE Trans Vis Comput Graph. 2018; 24:88–97.
Article54. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In : Advances in Neural Information Processing Systems. Proceedings of the 31st Neural Information Processing Systems (NIPS 2017); 2017 Dec 4-9; Long Beach, CA, USA. Red Hook: Curran Associates Inc;2017. 4765–74.55. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradientbased localization. Int J Comput Vision. 2019 Oct 11 [Epub]. https:// doi.org/10.1007/s11263-019-01228-7.
Article56. Ribeiro MT, Singh S, Guestrin C. Why should I trust you? Explaining the predictions of any classifier. New York: Association for Computing Machinery;2016. p. 1135–44.57. Ilse M, Tomczak JM, Welling M. Attention-based deep multiple instance learning. In : Proceedings of the 35th International Conference on Machine Learning; 2018 Jul 10-15; Stockholm, Sweden. Cambridge: Proceedings of Machine Learning Research;2018. 3376–91.58. Mothilal RK, Sharma A, Tan C. Explaining machine learning classifiers through diverse counterfactual explanations. Preprint at: https://arxiv.org/abs/1905.07697 (2019).
Article59. Richter MM, Weber RO. Case-based reasoning: a textbook. Berlin: Springer-Verlag;2013. p. 546.60. Watson I, Marir F. Case-based reasoning: a review. Knowl Eng Rev. 1994; 9:327–54.
Article61. Keane MT, Kenny EM. How case based reasoning explained neural networks: an XAI survey of post-hoc explanation-by-example in ANN-CBR twins. Preprint at: https://arxiv.org/abs/1905.07186 (2019).62. Hegde N, Hipp JD, Liu Y, et al. Similar image search for histopathology: SMILY. NPJ Digit Med. 2019; 2:56.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Digital Mammography
- Fabrication of additive manufacturing interim denture and comparison with conventional interim denture: A case report
- Fabricating a Ceramic-Pressed-to-Metal Restoration with Computer-Aided Design, Computer-Aided Manufacturing and Selective Laser Sintering: A Case Report
- Implant Restoration with Reverse Engineering and Computer-aided Design and Computer- aided Manufacturing Technology for a Discontinued Component: A Case Report
- Posterior rehabilitation considering mandibular movement with digital facebow transfer and virtual articulator: A case report