Korean Circ J.
2020 May;50(5):379-394. 10.4070/kcj.2019.0400.
Metabolic Crosstalk between the Heart and Fat
- Affiliations
-
- 1Center for Translational Medicine, Department of Pharmacology, Lewis Katz School of Medicine, Temple University, Philadelphia, PA, USA. walter.koch@temple.edu
- KMID: 2471777
- DOI: http://doi.org/10.4070/kcj.2019.0400
Abstract
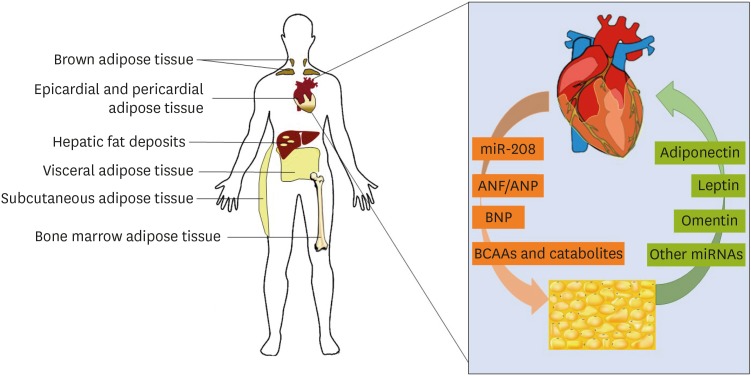
- It is now recognized that the heart can behave as a true endocrine organ, which can modulate the function of other tissues. Emerging evidence has shown that visceral fat is one such distant organ the heart communicates with. In fact, it appears that bi-directional crosstalk between adipose tissue and the myocardium is crucial to maintenance of normal function in both organs. In particular, factors secreted from the heart are now known to influence the metabolic activity of adipose tissue and other organs, as well as modulate the release of metabolic substrates and signaling molecules from the periphery. This review summarizes current knowledge regarding primary cardiokines and adipokines involved in heart-fat crosstalk, as well as implications of their dysregulation for cardiovascular health.
Keyword
MeSH Terms
Figure
Reference
-
1. Ogawa T, de Bold AJ. The heart as an endocrine organ. Endocr Connect. 2014; 3:R31–44. PMID: 24562677.2. de Bold AJ, Ma KK, Zhang Y, de Bold ML, Bensimon M, Khoshbaten A. The physiological and pathophysiological modulation of the endocrine function of the heart. Can J Physiol Pharmacol. 2001; 79:705–714. PMID: 11558679.3. Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005; 85:1093–1129. PMID: 15987803.4. Grynberg A, Demaison L. Fatty acid oxidation in the heart. J Cardiovasc Pharmacol. 1996; 28(Suppl 1):S11–7. PMID: 8891866.5. Collins S. A heart-adipose tissue connection in the regulation of energy metabolism. Nat Rev Endocrinol. 2014; 10:157–163. PMID: 24296515.6. Pascual F, Coleman RA. Fuel availability and fate in cardiac metabolism: a tale of two substrates. Biochim Biophys Acta. 2016; 1861:1425–1433. PMID: 26993579.7. Beatty CH, Young MK, Dwyer D, Bocek RM. Glucose utilization of cardiac and skeletal muscle homogenates from fetal and adult rhesus monkeys. Pediatr Res. 1972; 6:813–821. PMID: 4630128.8. Lopaschuk GD, Collins-Nakai RL, Itoi T. Developmental changes in energy substrate use by the heart. Cardiovasc Res. 1992; 26:1172–1180. PMID: 1288863.9. Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000; 106:847–856. PMID: 11018072.10. Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci. 2010; 1188:191–198. PMID: 20201903.11. Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007; 12:331–343. PMID: 17516164.12. Dirkx E, da Costa Martins PA, De Windt LJ. Regulation of fetal gene expression in heart failure. Biochim Biophys Acta. 2013; 1832:2414–2424. PMID: 24036209.13. Razeghi P, Young ME, Alcorn JL, Moravec CS, Frazier OH, Taegtmeyer H. Metabolic gene expression in fetal and failing human heart. Circulation. 2001; 104:2923–2931. PMID: 11739307.14. Chen CH, Liu YF, Lee SD, et al. Altitude hypoxia increases glucose uptake in human heart. High Alt Med Biol. 2009; 10:83–86. PMID: 19278356.15. Fewell JE, Zhang C. Heart glycogen influences protective responses of rat pups to hypoxia during early postnatal maturation. Biol Syst Open Access. 2013; 2:106.16. Handzlik MK, Constantin-Teodosiu D, Greenhaff PL, Cole MA. Increasing cardiac pyruvate dehydrogenase flux during chronic hypoxia improves acute hypoxic tolerance. J Physiol. 2018; 596:3357–3369. PMID: 29383727.17. Taegtmeyer H. Glycogen in the heart--an expanded view. J Mol Cell Cardiol. 2004; 37:7–10. PMID: 15242730.18. Sethi JK, Vidal-Puig AJ. Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007; 48:1253–1262. PMID: 17374880.19. Joyner JM, Hutley LJ, Cameron DP. Glucocorticoid receptors in human preadipocytes: regional and gender differences. J Endocrinol. 2000; 166:145–152. PMID: 10856893.20. Hellmér J, Marcus C, Sonnenfeld T, Arner P. Mechanisms for differences in lipolysis between human subcutaneous and omental fat cells. J Clin Endocrinol Metab. 1992; 75:15–20. PMID: 1320047.21. Imbeault P, Couillard C, Tremblay A, Després JP, Mauriège P. Reduced alpha(2)-adrenergic sensitivity of subcutaneous abdominal adipocytes as a modulator of fasting and postprandial triglyceride levels in men. J Lipid Res. 2000; 41:1367–1375. PMID: 10974043.22. Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008; 454:961–967. PMID: 18719582.23. Wilson-Fritch L, Burkart A, Bell G, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003; 23:1085–1094. PMID: 12529412.24. Bamshad M, Song CK, Bartness TJ. CNS origins of the sympathetic nervous system outflow to brown adipose tissue. Am J Physiol. 1999; 276:R1569–78. PMID: 10362733.25. Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. 1984; 167:10–14. PMID: 6698197.26. Rosenwald M, Wolfrum C. The origin and definition of brite versus white and classical brown adipocytes. Adipocyte. 2014; 3:4–9. PMID: 24575363.27. Boström P, Wu J, Jedrychowski MP, et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012; 481:463–468. PMID: 22237023.28. Liu D, Ceddia RP, Collins S. Cardiac natriuretic peptides promote adipose ‘browning’ through mTOR complex-1. Mol Metab. 2018; 9:192–198. PMID: 29396369.29. Sacks HS, Fain JN. Human epicardial adipose tissue: a review. Am Heart J. 2007; 153:907–917. PMID: 17540190.30. Iacobellis G, Corradi D, Sharma AM. Epicardial adipose tissue: anatomic, biomolecular and clinical relationships with the heart. Nat Clin Pract Cardiovasc Med. 2005; 2:536–543. PMID: 16186852.31. Antonopoulos AS, Margaritis M, Verheule S, et al. Mutual regulation of epicardial adipose tissue and myocardial redox state by PPAR-γ/adiponectin signalling. Circ Res. 2016; 118:842–855. PMID: 26838789.32. Antonopoulos AS, Antoniades C. The role of epicardial adipose tissue in cardiac biology: classic concepts and emerging roles. J Physiol. 2017; 595:3907–3917. PMID: 28191635.33. Graeff DB, Foppa M, Pires JC, et al. Epicardial fat thickness: distribution and association with diabetes mellitus, hypertension and the metabolic syndrome in the ELSA-Brasil study. Int J Cardiovasc Imaging. 2016; 32:563–572. PMID: 26585750.34. Iacobellis G. Epicardial fat thickness as a biomarker in cardiovascular disease. In : Patel VB, Preedy VR, editors. Biomarkers in Cardiovascular Disease. Dordrecht: Springer;2013. p. 1–11.35. Opincariu D, Mester A, Dobra M, Rat N, Hodas R, Morariu M. Prognostic value of epicardial fat thickness as a biomarker of increased inflammatory. status in patients with type 2 diabetes mellitus and acute myocardial infarction. J Cardiovasc Emerg. 2016; 2:11–18.36. Iacobellis G, Bianco AC. Epicardial adipose tissue: emerging physiological, pathophysiological and clinical features. Trends Endocrinol Metab. 2011; 22:450–457. PMID: 21852149.37. Münzberg H, Morrison CD. Structure, production and signaling of leptin. Metabolism. 2015; 64:13–23. PMID: 25305050.38. Schönke M, Björnholm M, Chibalin AV, Zierath JR, Deshmukh AS. Proteomics analysis of skeletal muscle from leptin-deficient ob/ob mice reveals adaptive remodeling of metabolic characteristics and fiber type composition. Proteomics. 2018; 18:e1700375. PMID: 29350465.39. Sáinz N, Barrenetxe J, Moreno-Aliaga MJ, Martínez JA. Leptin resistance and diet-induced obesity: central and peripheral actions of leptin. Metabolism. 2015; 64:35–46. PMID: 25497342.40. Puurunen VP, Kiviniemi A, Lepojärvi S, et al. Leptin predicts short-term major adverse cardiac events in patients with coronary artery disease. Ann Med. 2017; 49:448–454. PMID: 28300429.41. Hall ME, Harmancey R, Stec DE. Lean heart: role of leptin in cardiac hypertrophy and metabolism. World J Cardiol. 2015; 7:511–524. PMID: 26413228.42. Paolisso G, Tagliamonte MR, Galderisi M, et al. Plasma leptin level is associated with myocardial wall thickness in hypertensive insulin-resistant men. Hypertension. 1999; 34:1047–1052. PMID: 10567180.43. Dong F, Zhang X, Yang X, et al. Impaired cardiac contractile function in ventricular myocytes from leptin-deficient ob/ob obese mice. J Endocrinol. 2006; 188:25–36. PMID: 16394172.44. Purdham DM, Zou MX, Rajapurohitam V, Karmazyn M. Rat heart is a site of leptin production and action. Am J Physiol Heart Circ Physiol. 2004; 287:H2877–H2884. PMID: 15284063.45. Kadowaki T, Yamauchi T. Adiponectin and adiponectin receptors. Endocr Rev. 2005; 26:439–451. PMID: 15897298.46. Shapiro L, Scherer PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr Biol. 1998; 8:335–338. PMID: 9512423.47. Díez JJ, Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol. 2003; 148:293–300. PMID: 12611609.48. Arita Y, Kihara S, Ouchi N, et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999; 257:79–83. PMID: 10092513.49. Bauche IB, El Mkadem SA, Pottier AM, et al. Overexpression of adiponectin targeted to adipose tissue in transgenic mice: impaired adipocyte differentiation. Endocrinology. 2007; 148:1539–1549. PMID: 17204560.50. Rutter MK, Parise H, Benjamin EJ, et al. Impact of glucose intolerance and insulin resistance on cardiac structure and function: sex-related differences in the Framingham Heart Study. Circulation. 2003; 107:448–454. PMID: 12551870.51. Schannwell CM, Schneppenheim M, Perings S, Plehn G, Strauer BE. Left ventricular diastolic dysfunction as an early manifestation of diabetic cardiomyopathy. Cardiology. 2002; 98:33–39. PMID: 12373045.52. Guo Z, Xia Z, Yuen VG, McNeill JH. Cardiac expression of adiponectin and its receptors in streptozotocin-induced diabetic rats. Metabolism. 2007; 56:1363–1371. PMID: 17884446.53. Zhang L, Jaswal JS, Ussher JR, et al. Cardiac insulin-resistance and decreased mitochondrial energy production precede the development of systolic heart failure after pressure-overload hypertrophy. Circ Heart Fail. 2013; 6:1039–1048. PMID: 23861485.54. Jia G, Whaley-Connell A, Sowers JR. Diabetic cardiomyopathy: a hyperglycaemia- and insulin-resistance-induced heart disease. Diabetologia. 2018; 61:21–28. PMID: 28776083.55. Maeda N, Shimomura I, Kishida K, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002; 8:731–737. PMID: 12068289.56. Ohashi K, Parker JL, Ouchi N, et al. Adiponectin promotes macrophage polarization toward an anti-inflammatory phenotype. J Biol Chem. 2010; 285:6153–6160. PMID: 20028977.57. Holland WL, Miller RA, Wang ZV, et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med. 2011; 17:55–63. PMID: 21186369.58. Fain JN, Sacks HS, Buehrer B, et al. Identification of omentin mRNA in human epicardial adipose tissue: comparison to omentin in subcutaneous, internal mammary artery periadventitial and visceral abdominal depots. Int J Obes. 2008; 32:810–815.59. de Souza Batista CM, Yang RZ, Lee MJ, et al. Omentin plasma levels and gene expression are decreased in obesity. Diabetes. 2007; 56:1655–1661. PMID: 17329619.60. Matsuo K, Shibata R, Ohashi K, et al. Omentin functions to attenuate cardiac hypertrophic response. J Mol Cell Cardiol. 2015; 79:195–202. PMID: 25479337.61. Kutlay Ö, Kaygısız Z, Kaygısız B. Effect of omentin on cardiovascular functions and gene expressions in isolated rat hearts. Anatol J Cardiol. 2019; 21:91–97. PMID: 30694801.62. Kataoka Y, Shibata R, Ohashi K, et al. Omentin prevents myocardial ischemic injury through AMP-activated protein kinase- and Akt-dependent mechanisms. J Am Coll Cardiol. 2014; 63:2722–2733. PMID: 24768874.63. Yamawaki H, Tsubaki N, Mukohda M, Okada M, Hara Y. Omentin, a novel adipokine, induces vasodilation in rat isolated blood vessels. Biochem Biophys Res Commun. 2010; 393:668–672. PMID: 20170632.64. Kazama K, Okada M, Hara Y, Yamawaki H. A novel adipocytokine, omentin, inhibits agonists-induced increases of blood pressure in rats. J Vet Med Sci. 2013; 75:1029–1034. PMID: 23546685.65. Herman MA, She P, Peroni OD, Lynch CJ, Kahn BB. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. J Biol Chem. 2010; 285:11348–11356. PMID: 20093359.66. Grajeda-Iglesias C, Aviram M. Specific amino acids affect cardiovascular diseases and atherogenesis via protection against macrophage foam cell formation: review article. Rambam Maimonides Med J. 2018; 9:e0022.67. Sun H, Olson KC, Gao C, et al. Catabolic defect of branched-chain amino acids promotes heart failure. Circulation. 2016; 133:2038–2049. PMID: 27059949.68. Ciccarelli M, Chuprun JK, Rengo G, et al. G protein-coupled receptor kinase 2 activity impairs cardiac glucose uptake and promotes insulin resistance after myocardial ischemia. Circulation. 2011; 123:1953–1962. PMID: 21518983.69. Woodall BP, Gresham KS, Woodall MA, et al. Alteration of myocardial GRK2 produces a global metabolic phenotype. JCI insight. 2019; 5:123848. PMID: 30946029.70. Sato P, Chuprun JK, Grisanti L, et al. GRK2-S670A mice reveal cardioprotection post ischemia-reperfusion. J Mol Cell Cardiol. 2017; 112:152–153.71. Chua B, Siehl DL, Morgan HE. Effect of leucine and metabolites of branched chain amino acids on protein turnover in heart. J Biol Chem. 1979; 254:8358–8362. PMID: 468830.72. Ruiz-Canela M, Toledo E, Clish CB, et al. Plasma branched-chain amino acids and incident cardiovascular disease in the PREDIMED trial. Clin Chem. 2016; 62:582–592. PMID: 26888892.73. Li T, Zhang Z, Kolwicz SC Jr, et al. Defective branched-chain amino acid catabolism disrupts glucose metabolism and sensitizes the heart to ischemia-reperfusion injury. Cell Metab. 2017; 25:374–385. PMID: 28178567.74. Green CR, Wallace M, Divakaruni AS, et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat Chem Biol. 2016; 12:15–21. PMID: 26571352.75. Halama A, Horsch M, Kastenmüller G, et al. Metabolic switch during adipogenesis: From branched chain amino acid catabolism to lipid synthesis. Arch Biochem Biophys. 2016; 589:93–107. PMID: 26408941.76. Neinast MD, Jang C, Hui S, et al. Quantitative analysis of the whole-body metabolic fate of branched-chain amino acids. Cell Metab. 2019; 29:417–429.e4. PMID: 30449684.77. Jamieson JD, Palade GE. Specific granules in atrial muscle cells. J Cell Biol. 1964; 23:151–172. PMID: 14228508.78. Baines AD, DeBold AJ, Sonnenberg H. Natriuretic effect of atrial extract on isolated perfused rat kidney. Can J Physiol Pharmacol. 1983; 61:1462–1466. PMID: 6671158.79. Saito Y, Nakao K, Itoh H, et al. Brain natriuretic peptide is a novel cardiac hormone. Biochem Biophys Res Commun. 1989; 158:360–368. PMID: 2521788.80. Hosoda K, Nakao K, Mukoyama M. Expression of brain natriuretic peptide gene in human heart. Production in the ventricle. Hypertension. 1991; 17:1152–1155. PMID: 2045161.81. Dewey CM, Spitler KM, Ponce JM, Hall DD, Grueter CE. Cardiac-secreted factors as peripheral metabolic regulators and potential disease biomarkers. J Am Heart Assoc. 2016; 5:e003101. PMID: 27247337.82. Wang TJ, Larson MG, Levy D, et al. Impact of obesity on plasma natriuretic peptide levels. Circulation. 2004; 109:594–600. PMID: 14769680.83. Plante E, Menaouar A, Danalache BA, Broderick TL, Jankowski M, Gutkowska J. Treatment with brain natriuretic peptide prevents the development of cardiac dysfunction in obese diabetic db/db mice. Diabetologia. 2014; 57:1257–1267. PMID: 24595856.84. Cabiati M, Raucci S, Liistro T, et al. Impact of obesity on the expression profile of natriuretic peptide system in a rat experimental model. PLoS One. 2013; 8:e72959. PMID: 24009719.85. Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006; 27:47–72. PMID: 16291870.86. Sarzani R, Dessì-Fulgheri P, Paci VM, Espinosa E, Rappelli A. Expression of natriuretic peptide receptors in human adipose and other tissues. J Endocrinol Invest. 1996; 19:581–585. PMID: 8957740.87. Sengenès C, Berlan M, De Glisezinski I, Lafontan M, Galitzky J. Natriuretic peptides: a new lipolytic pathway in human adipocytes. FASEB J. 2000; 14:1345–1351. PMID: 10877827.88. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015; 87:3–14. PMID: 25979468.89. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005; 436:214–220. PMID: 15951802.90. Fernández-Hernando C, Suárez Y, Rayner KJ, Moore KJ. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011; 22:86–92. PMID: 21178770.91. Kim SY, Kim AY, Lee HW, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010; 392:323–328. PMID: 20060380.92. Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009; 58:1050–1057. PMID: 19188425.93. Ahn J, Lee H, Jung CH, Jeon TI, Ha TY. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO Mol Med. 2013; 5:1602–1612. PMID: 24009212.94. Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011; 48:61–69. PMID: 20857148.95. Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017; 542:450–455. PMID: 28199304.96. Iacomino G, Russo P, Stillitano I, et al. Circulating microRNAs are deregulated in overweight/obese children: preliminary results of the I.Family study. Genes Nutr. 2016; 11:7. PMID: 27551310.97. Boštjančič E, Zidar N, Štajer D, Glavač D. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology. 2010; 115:163–169. PMID: 20029200.98. van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006; 103:18255–18260. PMID: 17108080.99. van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007; 117:2369–2376. PMID: 17786230.100. Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009; 119:2772–2786. PMID: 19726871.101. Grueter CE, van Rooij E, Johnson BA, et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012; 149:671–683. PMID: 22541436.102. Pospisilik JA, Schramek D, Schnidar H, et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell. 2010; 140:148–160. PMID: 20074523.103. Baskin KK, Grueter CE, Kusminski CM, et al. MED13-dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. EMBO Mol Med. 2014; 6:1610–1621. PMID: 25422356.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Extracellular Mechanisms of Neutrophils in Immune Cell Crosstalk
- Relationship Between Pulmonary Function and Metabolic Syndrome, Body Mass Index, Fat Percentage, and Fat Mass
- Exploring the Crosstalk between Adipose Tissue and the Cardiovascular System
- Relationship between Metabolic Syndrome and Coronary Heart Disease in Elderly
- Being Metabolically Healthy, the Most Responsible Factor for Vascular Health