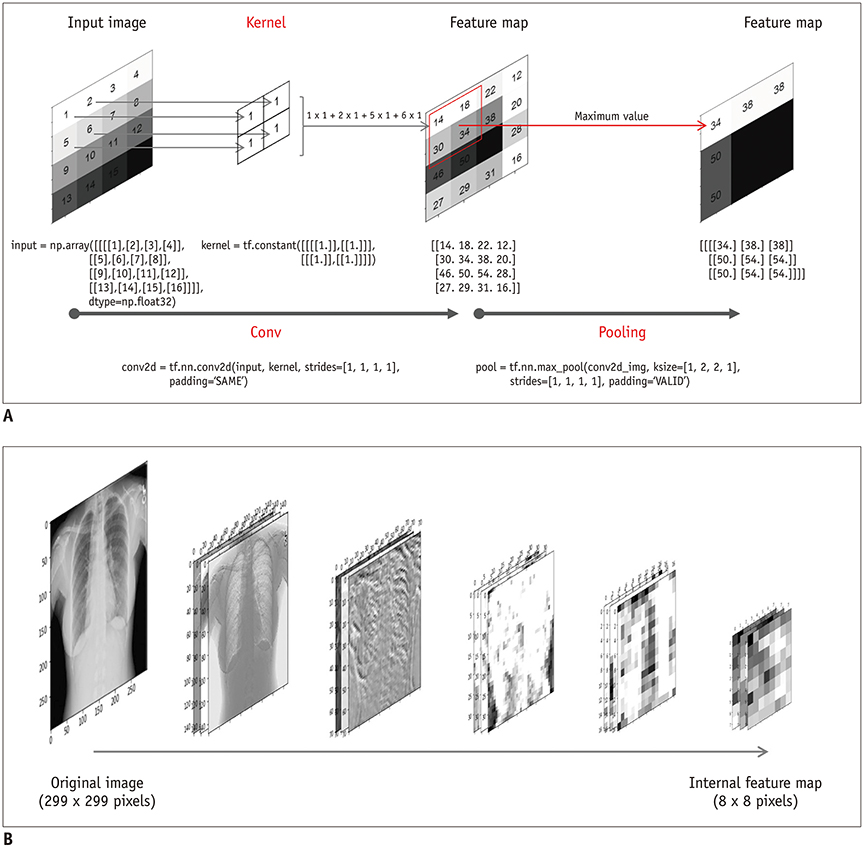
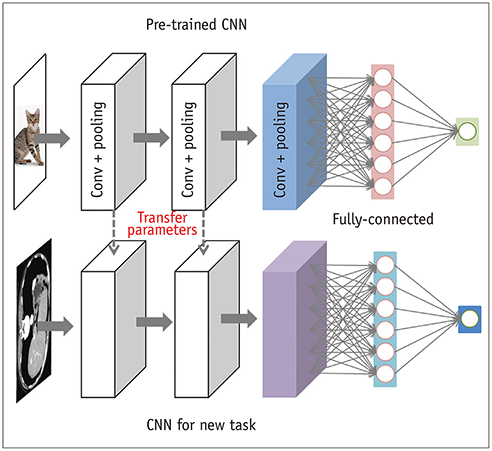
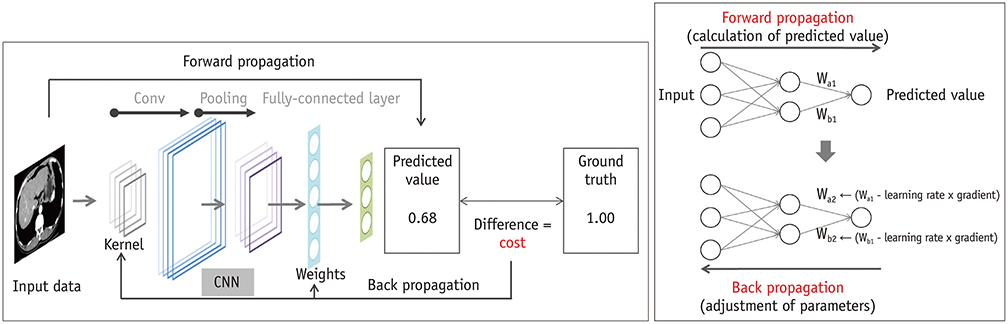
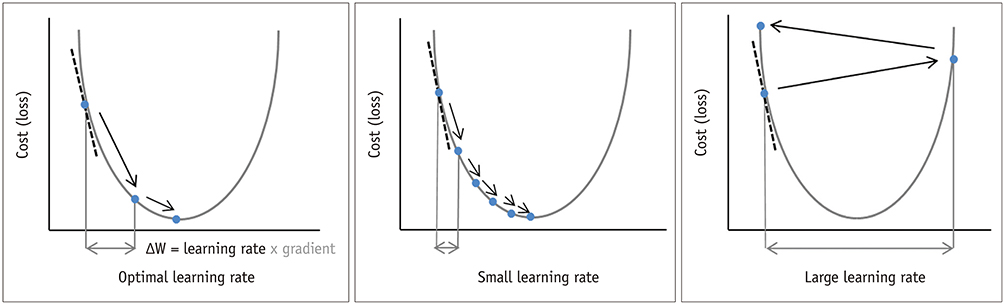
Korean J Radiol.
2020 Jan;21(1):33-41. 10.3348/kjr.2019.0312.
Basics of Deep Learning: A Radiologist's Guide to Understanding Published Radiology Articles on Deep Learning
- Affiliations
-
- 1Department of Radiology, Massachusetts General Hospital, Boston, MA, USA. kdsong0308@gmail.com
- 2Department of Radiology, Sungkyunkwan University School of Medicine, Samsung Medical Center, Seoul, Korea.
- 3Department of Internal Medicine, National Medical Center, Seoul, Korea.
- KMID: 2467040
- DOI: http://doi.org/10.3348/kjr.2019.0312
Abstract
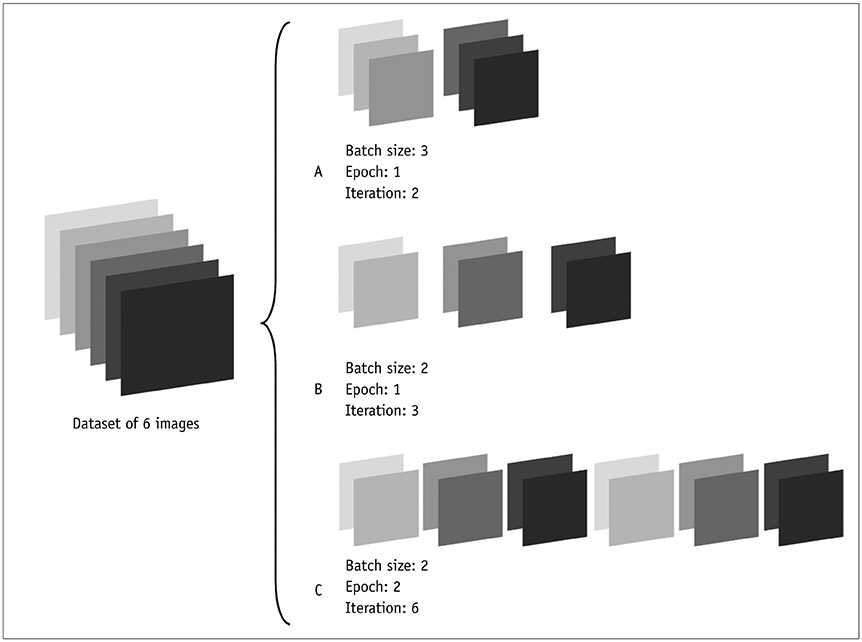
- Artificial intelligence has been applied to many industries, including medicine. Among the various techniques in artificial intelligence, deep learning has attained the highest popularity in medical imaging in recent years. Many articles on deep learning have been published in radiologic journals. However, radiologists may have difficulty in understanding and interpreting these studies because the study methods of deep learning differ from those of traditional radiology. This review article aims to explain the concepts and terms that are frequently used in deep learning radiology articles, facilitating general radiologists' understanding.
Figure
Reference
-
1. Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics. 2017; 37:2113–2131.
Article2. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017; 42:60–88.
Article3. Dreyer KJ, Geis JR. When machines think: radiology’s next frontier. Radiology. 2017; 285:713–718.
Article4. Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional neural networks for radiologic images: a radiologist's guide. Radiology. 2019; 290:590–606.
Article5. Cardoso JR, Pereira LM, Iversen MD, Ramos AL. What is gold standard and what is ground truth? Dental Press J Orthod. 2014; 19:27–30.
Article6. Zhong Z, Zheng L, Kang G, Li S, Yang Y. Random erasing data augmentation. eprint arXiv;2017. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2017arXiv170804896Z.7. TIOBE index for April 2019. TIOBE Web site;Accessed April 30, 2019. https://www.tiobe.com/tiobe-index/.8. Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, et al. Caffe: convolutional architecture for fast feature embedding. eprint arXiv;2014. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2014arXiv1408.5093J.9. Bastien F, Lamblin P, Pascanu R, Bergstra J, Goodfellow I, Bergeron A, et al. Theano: new features and speed improvements. eprint arXiv;2012. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2012arXiv1211.5590B.10. Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. TensorFlow: a system for large-scale machine learning. eprint arXiv;2016. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2016arXiv160508695A.11. Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017; 18:570–584.
Article12. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012; 25:1090–1098.
Article13. Lee C, Kim Y, Kim YS, Jang J. Automatic disease annotation from radiology reports using artificial intelligence implemented by a recurrent neural network. AJR Am J Roentgenol. 2019; 212:734–740.
Article14. Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. eprint arXiv;2013. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2013arXiv1311.2524G.15. Kazuhiro K, Werner RA, Toriumi F, Javadi MS, Pomper MG, Solnes LB, et al. Generative adversarial networks for the creation of realistic artificial brain magnetic resonance images. Tomography. 2018; 4:159–163.
Article16. Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. eprint arXiv;2014. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2014arXiv1406.2661G.17. Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. eprint arXiv;2015. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2015arXiv150504597R.18. Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. eprint arXiv;2016. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2016arXiv160806993H.19. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. eprint arXiv;2014. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2014arXiv1409.1556S.20. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. eprint arXiv;2015. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2015arXiv151203385H.21. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. eprint arXiv;2014. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2014arXiv1409.4842S.22. He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. eprint arXiv;2017. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2017arXiv170306870H.23. Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proceedings of the IEEE. 1998; 86:2278–2324.
Article24. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. eprint arXiv;2013. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2013arXiv1311.2901Z.25. Qian N. On the momentum term in gradient descent learning algorithms. Neural Netw. 1999; 12:145–151.
Article26. Nesterov YE. A method for solving the convex programming problem with convergence rate O(1/k2). Dokl Akad Nauk SSSR. 1983; 269:543–547.27. Duchi J, Hazan E, Singer Y. Adaptive subgradient methods for online learning and stochastic optimization. J Mach Learn Res. 2011; 12:2121–2159.28. Zeiler MD. ADADELTA: an adaptive learning rate method. eprint arXiv;2012. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2012arXiv1212.5701Z.29. Kingma DP, Ba J. Adam: a method for stochastic optimization. eprint arXiv;2014. Accessed April 1, 2019. Available at: https://ui.adsabs.harvard.edu/abs/2014arXiv1412.6980K.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current status of deep learning applications in abdominal ultrasonography
- Development of an Optimized Deep Learning Model for Medical Imaging
- An overview of deep learning in the field of dentistry
- Deep Learning in Dental Radiographic Imaging
- Deep Learning in MR Motion Correction:a Brief Review and a New Motion Simulation Tool (view2Dmotion)