Korean J Radiol.
2019 Jul;20(7):1124-1137. 10.3348/kjr.2018.0070.
Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives
- Affiliations
-
- 1Department of Radiology and Research Institute of Radiology, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea.
- 2Department of Clinical Epidemiology and Biostatistics, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea. hello.hello.hj@gmail.com
- KMID: 2467023
- DOI: http://doi.org/10.3348/kjr.2018.0070
Abstract
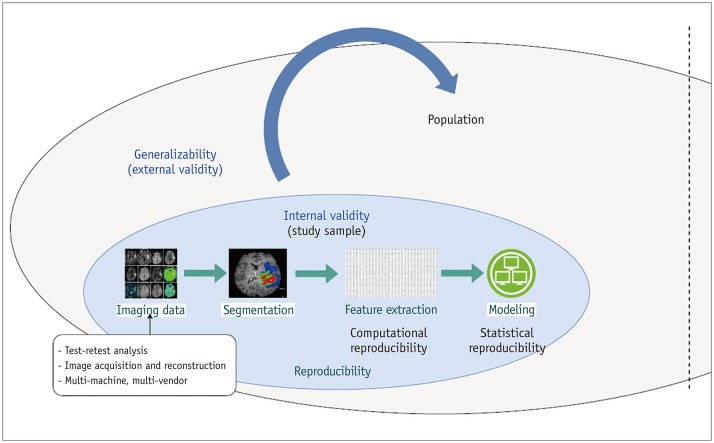
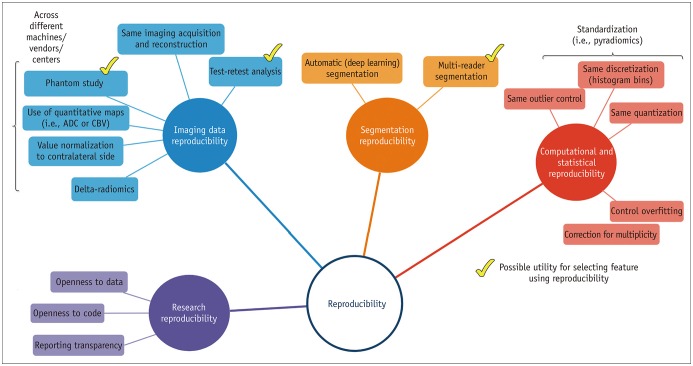
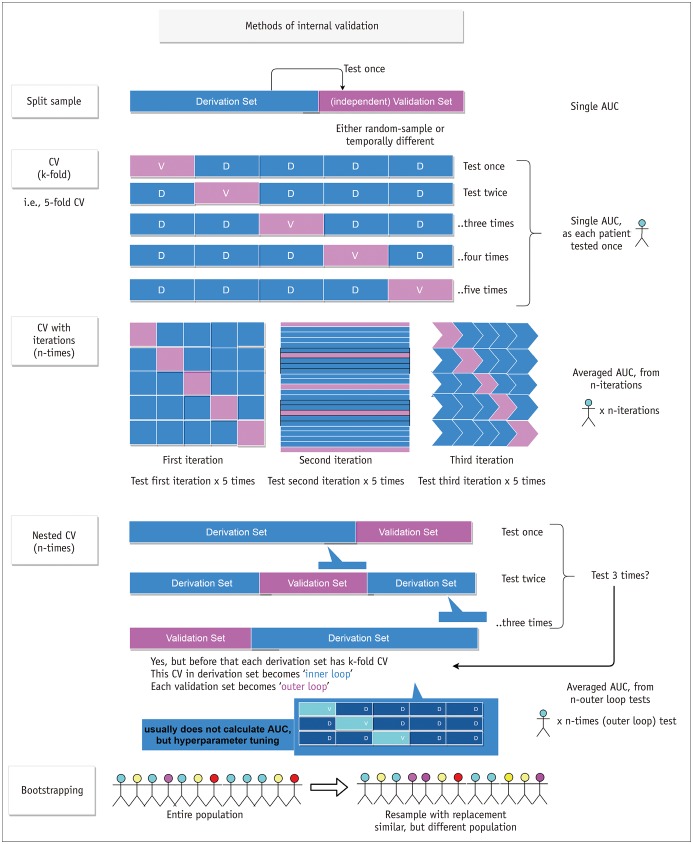
- Radiomics, which involves the use of high-dimensional quantitative imaging features for predictive purposes, is a powerful tool for developing and testing medical hypotheses. Radiologic and statistical challenges in radiomics include those related to the reproducibility of imaging data, control of overfitting due to high dimensionality, and the generalizability of modeling. The aims of this review article are to clarify the distinctions between radiomics features and other omics and imaging data, to describe the challenges and potential strategies in reproducibility and feature selection, and to reveal the epidemiological background of modeling, thereby facilitating and promoting more reproducible and generalizable radiomics research.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Radiomics and Deep Learning: Hepatic Applications
Hyo Jung Park, Bumwoo Park, Seung Soo Lee
Korean J Radiol. 2020;21(4):387-401. doi: 10.3348/kjr.2019.0752.
Reference
-
1. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016; 278:563–577. PMID: 26579733.
Article2. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012; 48:441–446. PMID: 22257792.
Article3. Micheel CM, Nass SJ, Omenn GS. Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials. Board on Health Care Services. Board on Health Sciences Policy. Institute of Medicine. Evolution of translational omics: lessons learned and the path forward. Washington, DC: The National Academies Press;2012.4. Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008; 105:5213–5218. PMID: 18362333.
Article5. Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007; 25:675–680. PMID: 17515910.
Article6. Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016; 61:R150–R166. PMID: 27269645.
Article7. Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and promises of PET radiomics. Int J Radiat Oncol Biol Phys. 2018; 102:1083–1089. PMID: 29395627.
Article8. Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017; 28:1191–1206. PMID: 28168275.
Article9. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012; 30:1234–1248. PMID: 22898692.
Article10. Park JE, Kim HS. Radiomics as a quantitative imaging biomarker: practical considerations and the current standpoint in neuro-oncologic studies. Nucl Med Mol Imaging. 2018; 52:99–108. PMID: 29662558.
Article11. Jain R, Lui YW. How far are we from using radiomics assessment of gliomas in clinical practice? Radiology. 2018; 289:807–808. PMID: 30277444.
Article12. Lee G, Lee HY, Park H, Schiebler ML, van Beek EJR, Ohno Y, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur J Radiol. 2017; 86:297–307. PMID: 27638103.
Article13. Kristensen VN, Lingjærde OC, Russnes HG, Vollan HK, Frigessi A, Børresen-Dale AL. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer. 2014; 14:299–313. PMID: 24759209.
Article14. Clarke R, Ressom HW, Wang A, Xuan J, Liu MC, Gehan EA, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008; 8:37–49. PMID: 18097463.
Article15. Jain AK, Duin RP, Mao J. Statistical pattern recognition: a review. IEEE Trans Pattern Anal Mach Intell. 2000; 22:4–37.
Article16. Bellman RE. Adaptive control processes: a guided tour. Princeton, NJ: Princeton university press;2015.17. Ferté C, Trister AD, Huang E, Bot BM, Guinney J, Commo F, et al. Impact of bioinformatic procedures in the development and translation of high-throughput molecular classifiers in oncology. Clin Cancer Res. 2013; 19:4315–4325. PMID: 23780890.
Article18. Genders TS, Spronk S, Stijnen T, Steyerberg EW, Lesaffre E, Hunink MG. Methods for calculating sensitivity and specificity of clustered data: a tutorial. Radiology. 2012; 265:910–916. PMID: 23093680.
Article19. Lin YC, Lin G, Hong JH, Lin YP, Chen FH, Ng SH, et al. Diffusion radiomics analysis of intratumoral heterogeneity in a murine prostate cancer model following radiotherapy: pixelwise correlation with histology. J Magn Reson Imaging. 2017; 46:483–489. PMID: 28176411.
Article20. O'Connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017; 14:169–186. PMID: 27725679.21. Zinn PO, Singh SK, Kotrotsou A, Hassan I, Thomas G, Luedi MM, et al. A coclinical radiogenomic validation study: conserved magnetic resonance radiomic appearance of periostin-expressing glioblastoma in patients and xenograft models. Clin Cancer Res. 2018; 24:6288–6299. PMID: 30054278.
Article22. Cook TD, Campbell DT. Quasi-experimentation: design & analysis issues for field settings. Chicago, IL: Rand McNally;1979.23. Ferguson L. External validity, generalizability, and knowledge utilization. J Nurs Scholarsh. 2004; 36:16–22. PMID: 15098414.
Article24. Murad MH, Katabi A, Benkhadra R, Montori VM. External validity, generalisability, applicability and directness: a brief primer. BMJ Evid Based Med. 2018; 23:17–19.
Article25. Raunig DL, McShane LM, Pennello G, Gatsonis C, Carson PL, Voyvodic JT, et al. ; QIBA Technical Performance Working Group. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res. 2015; 24:27–67. PMID: 24919831.26. Kessler LG, Barnhart HX, Buckler AJ, Choudhury KR, Kondratovich MV, Toledano A, et al. ; QIBA Terminology Working Group. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat Methods Med Res. 2015; 24:9–26. PMID: 24919826.27. Stodden V, Guo P, Ma Z. Toward reproducible computational research: an empirical analysis of data and code policy adoption by journals. PLoS One. 2013; 8:e67111. PMID: 23805293.
Article28. Stodden V, Leisch F, Peng RD. Implementing reproducible research. 1st ed. Boca Raton, FL: CRC Press;2014.29. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014; 5:4006. PMID: 24892406.
Article30. Antunes J, Viswanath S, Rusu M, Valls L, Hoimes C, Avril N, et al. Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: a proof-of-concept study. Transl Oncol. 2016; 9:155–162. PMID: 27084432.
Article31. Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014; 27:805–823. PMID: 24990346.
Article32. Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014; 273:168–174. PMID: 24827998.
Article33. Hunter LA, Krafft S, Stingo F, Choi H, Martel MK, Kry SF, et al. High quality machine-robust image features: identification in nonsmall cell lung cancer computed tomography images. Med Phys. 2013; 40:121916. PMID: 24320527.
Article34. Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, et al. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013; 52:1391–1397. PMID: 24047337.
Article35. Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. 2012; 53:693–700. PMID: 22454484.
Article36. van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, et al. Repeatability of radiomic features in non-small-cell lung cancer [(18)F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016; 18:788–795. PMID: 26920355.
Article37. van Velden FH, Nissen IA, Jongsma F, Velasquez LM, Hayes W, Lammertsma AA, et al. Test-retest variability of various quantitative measures to characterize tracer uptake and/or tracer uptake heterogeneity in metastasized liver for patients with colorectal carcinoma. Mol Imaging Biol. 2014; 16:13–18. PMID: 23807457.
Article38. Zhao B, James LP, Moskowitz CS, Guo P, Ginsberg MS, Lefkowitz RA, et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology. 2009; 252:263–272. PMID: 19561260.
Article39. Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015; 50:757–765. PMID: 26115366.
Article40. van Timmeren JE, Leijenaar RTH, van Elmpt W, Wang J, Zhang Z, Dekker A, et al. Test-retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography. 2016; 2:361–365. PMID: 30042967.
Article41. Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018; 288:407–415. PMID: 29688159.42. Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010; 49:1012–1016. PMID: 20831489.
Article43. He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016; 6:34921. PMID: 27721474.
Article44. Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One. 2016; 11:e0166550. PMID: 28033372.
Article45. Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Götz M, Wick A, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018; 20:848–857. PMID: 29036412.
Article46. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014; 9:e102107. PMID: 25025374.
Article47. Pavic M, Bogowicz M, Würms X, Glatz S, Finazzi T, Riesterer O, et al. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018; 57:1070–1074. PMID: 29513054.
Article48. Bogowicz M, Riesterer O, Bundschuh RA, Veit-Haibach P, Hüllner M, Studer G, et al. Stability of radiomic features in CT perfusion maps. Phys Med Biol. 2016; 61:8736–8749. PMID: 27893446.
Article49. Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, et al. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep. 2017; 7:14331. PMID: 29085044.
Article50. Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A, et al. Robustness of radiomic features in [11C]choline and [18F] FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol. 2016; 18:935–945. PMID: 27324369.51. Traverso A, Wee L, Dekker A, Gillies R. EP-2132: repeatability and reproducibility of radiomic features: results of a systematic review. Radiother Oncol. 2018; 127:S1174–S1175.
Article52. Hatt M, Majdoub M, Vallières M, Tixier F, Le Rest CC, Groheux D, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med. 2015; 56:38–44. PMID: 25500829.
Article53. Kang D, Park JE, Kim YH, Kim JH, Oh JY, Kim J, et al. Diffusion radiomics as a diagnostic model for atypical manifestation of primary central nervous system lymphoma: development and multicenter external validation. Neuro Oncol. 2018; 20:1251–1261. PMID: 29438500.
Article54. Kim JY, Park JE, Jo Y, Shim WH, Nam SJ, Kim JH, et al. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol. 2019; 21:404–414. PMID: 30107606.
Article55. Beukinga RJ, Hulshoff JB, Mul VEM, Noordzij W, Kats-Ugurlu G, Slart RHJA, et al. Prediction of response to neoadjuvant chemotherapy and radiation therapy with baseline and restaging 18F-FDG PET imaging biomarkers in patients with esophageal cancer. Radiology. 2018; 287:983–992. PMID: 29533721.56. Boldrini L, Cusumano D, Chiloiro G, Casà C, Masciocchi C, Lenkowicz J, et al. Delta radiomics for rectal cancer response prediction with hybrid 0.35 T magnetic resonance-guided radiotherapy (MRgRT): a hypothesis-generating study for an innovative personalized medicine approach. Radiol Med. 2019; 124:145–153. PMID: 30374650.57. Crombé A, Périer C, Kind M, De Senneville BD, Le Loarer F, Italiano A, et al. T2-based MRI delta-radiomics improve response prediction in soft-tissue sarcomas treated by neoadjuvant chemotherapy. J Magn Reson Imaging. 2018; 12:577. DOI: 10.1002/jmri.26589. [Epub ahead of print].58. Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep. 2017; 7:588. PMID: 28373718.
Article59. Mazzei MA, Nardone V, Di Giacomo L, Bagnacci G, Gentili F, Tini P, et al. The role of delta radiomics in gastric cancer. Quant Imaging Med Surg. 2018; 8:719–721. PMID: 30211038.
Article60. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017; 14:749–762. PMID: 28975929.
Article61. Doumou G, Siddique M, Tsoumpas C, Goh V, Cook GJ. The precision of textural analysis in (18)F-FDG-PET scans of oesophageal cancer. Eur Radiol. 2015; 25:2805–2812. PMID: 25994189.
Article62. Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with Deep Neural Networks. Med Image Anal. 2017; 35:18–31. PMID: 27310171.
Article63. Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015; 34:1993–2024. PMID: 25494501.
Article64. van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017; 77:e104–e107. PMID: 29092951.
Article65. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8:118–127. PMID: 16632515.
Article66. Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018; 59:1321–1328. PMID: 29301932.
Article67. Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology. 2019; 291:53–59. PMID: 30694160.
Article68. Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007; 23:2507–2517. PMID: 17720704.
Article69. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995; 57:289–300.
Article70. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001; 98:5116–5121. PMID: 11309499.
Article71. Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003; 19:368–375. PMID: 12584122.
Article72. Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 2003; 31:2013–2035.
Article73. Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. Berlin/Heidelberg: Springer Science & Business Media;2008.74. Shmueli G. To explain or to predict? Statistical Science. 2010; 25:289–310.
Article75. Gordis L. Assessing the validity and reliability of diagnostic and screening tests. In : Gordis L, editor. Epidemiology. 5th ed. London: Elsevier Health Sciences;2013.76. Park SH. Diagnostic case-control versus diagnostic cohort studies for clinical validation of artificial intelligence algorithm performance. Radiology. 2019; 290:272–273. PMID: 30511912.
Article77. Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018; 286:800–809. PMID: 29309734.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Radiomics in Oncological PET/CT: a Methodological Overview
- Radiomics as a Quantitative Imaging Biomarker: Practical Considerations and the Current Standpoint in Neuro-oncologic Studies
- A Tale of Two Fields: Mathematical and Statistical Modeling of Infectious Diseases
- A Factor Analysis of the Perspectives on the Coping Strategies about Practical Stress in Nursing Student
- Radiomics in Breast Imaging from Techniques to Clinical Applications: A Review